Molecular Definition Implementation Guide for Molecular Data Types, published by HL7 International / Clinical Genomics. This guide is not an authorized publication; it is the continuous build for version 1.0.0-ballot1 built by the FHIR (HL7® FHIR® Standard) CI Build. This version is based on the current content of https://github.com/HL7/molecular-definition-data-types/ and changes regularly. See the Directory of published versions
| Official URL: http://hl7.org/fhir/uv/molecular-definition-data-types/ImplementationGuide/hl7.fhir.uv.molecular-definition-data-types | Version: 1.0.0-ballot1 | |||
| IG Standards status: Draft | Maturity Level: 0 | Computable Name: MolecularDefinitionDataTypes | ||
Structured genetic information plays a crucial role in modern genomic medicine, enabling precision-based approaches such as personalized medicine and targeted therapies. Accurate representation and exchange of genetic data are essential for clinical decision-making, research, and interoperability across healthcare systems. However, the inherent complexity of genetic definitions, along with variations in interpretation based on context and use case, poses significant challenges in standardizing genetic data communication.
This implementation guide is not complete. The included artifacts are marked as experimental, but they are ready for review, testing, and validation.
This implementation guide addresses these challenges by leveraging the newly developed MolecularDefinition resource to support the structured representation of fundamental genetic concepts, including sequence, allele, and variation. Additionally, it integrates various core FHIR resources to establish a robust framework for interoperable genetic data exchange.
The guide outlines methodologies and provides practical examples of how to utilize the developed MolecularDefinition profiles across diverse genomic applications, including:
By establishing a standardized approach to genomic data representation, this implementation guide facilitates consistent and accurate genetic information exchange, supporting integration within electronic health records, research databases, and clinical genomic workflows
This Implementation Guide is designed to assist system developers and standards implementers in adopting the FHIR genomic profiles of the MolecularDefinition resource across diverse use cases. The implementation guide defines five key profiles—Sequence, Allele, Variation, Haplotype, and Genotype—each tailored to represent distinct genomic concepts. To ensure consistent and reliable representation of the various data elements within these profiles, the guide also provides a curated set of Code Systems and Value Sets. These terminologies, currently bound with example strength to their respective elements, are expected to evolve and undergo validation over time, while already supporting meaningful and accurate data representation. The included Code Systems and Value Sets comprise a combination of adapted existing ontologies and terminologies alongside newly developed vocabularies specifically crafted for the MolecularDefinition structure. Furthermore, the Use Cases section illustrates practical applications of these MolecularDefinition profiles, Code Systems, and Value Sets through real-world genomic molecule examples, facilitating a deeper understanding of their implementation in clinical and research contexts
Readers are encouraged to start with this page and follow the linked sections in order. They are also free to navigate directly to any section of interest or move back and forth according to their individual needs—for example, jumping directly to the Use Case section. The recommended reading sequence starts with the Motivation section, which provides the foundational rationale behind the MolecularDefinition profiles. Following this, readers should familiarize themselves with the MolecularDefinition core resource to understand its structure and scope. Next, it is important to grasp the intended goals of each profile, including their distinctions and interrelationships. Afterward, an overview of the curated Code Systems should be reviewed to appreciate their specific purposes and how they support the corresponding profiles. Finally, readers may explore the practical examples beginning with simpler Sequence, Allele, and Variation example instances, progressively advancing to the more complex scenarios involving Haplotypes and Genotypes.
Suggested Sequence:
This Implementation Guide is designed to define foundational genomic elements of genomic molecules and concepts—such as sequences and alleles—adopting a patient-agnostic perspective crucial for establishing standardized genomic FHIR artifacts. The Clinical Genomics Working Group also supports the Genomics Reporting Implementation Guide (GRIG) which includes concepts like the genomic report itself and the high-level categories of observations and other elements that make up the report.
As of this release, this IG and the GRIG are independent of each other. This IG is ready for testing its intended goals of representing the foundational definitional elements. This IG is intended to support any workflow where there is a need for explicit description of the concepts it defines. All of the profiles defined here will be patient-agnostic, so if your use case calls for reporting patient specific results, we would recommend utilizing the GRIG for now.
In the future, the Working Group will utilize the discrete data structures defined by this IG within the GRIG, by addition to and/or substitution of corresponding GRIG profiles (e.g., Variant, Genotype, Haplotype, etc …). We anticipate an improved ability to exchange structured genomic data produced by clinical testing. For example, it is possible the existing GRIG profiles will remain for the community, and a new set of Observation profiles utilizing this IG will be added to the GRIG. Alternatively, the existing profiles in the GRIG might be redesigned to use this IG. The value of this IG is in the loose coupling between the well-defined and discrete data structures, so ultimately this IG will add value to genomic data interoperability regardless of the decisions or timelines related to its inclusion within the GRIG.
This diagram is the current strategic vision, which is still a work in progress and subject to change. As with all our work, these decisions will be subject to open discussion and formal votes. Timelines for implementation will be dependent on the feedback and testing.
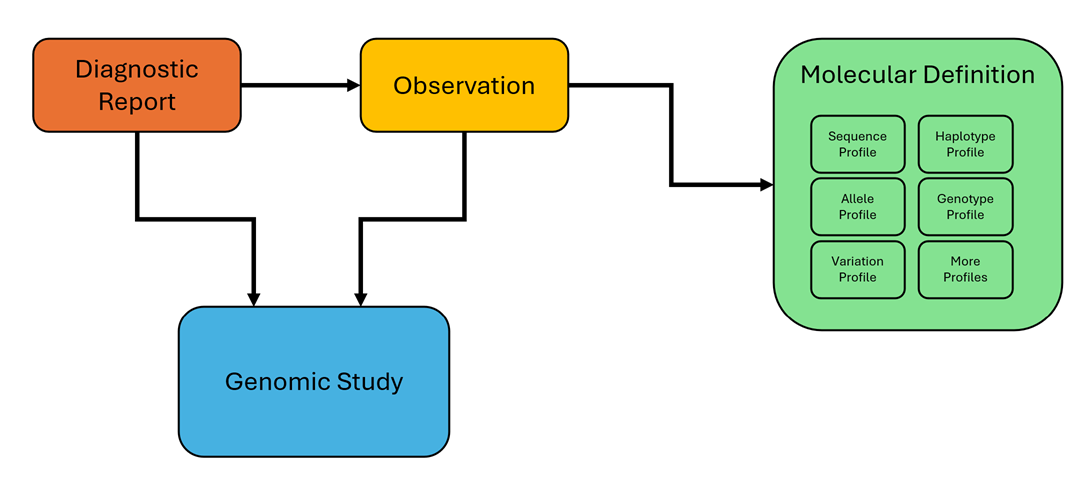
The Sequence profile is a core part of this guide, used to represent raw genetic sequences like DNA, RNA, and proteins in various formats. Unlike profiles such as Allele and Variation that emphasize genomic location, the Sequence profile focuses on the intrinsic composition of genetic material and supports integration with other profiles to enhance overall functionality. The Allele profile builds upon the Sequence profile by defining genetic makeup at specific genomic locations through two states: the base context and the specific allele state. This structured representation ensures accuracy and consistency, forming a basis for other profiles to leverage. The Variation profile represents genetic variations at a precise location, characterizing genetic composition in three states—context, reference, and alternative—while enabling integration with Sequence profile for comprehensive depiction of genomic alterations. The Haplotype profile captures collections of related alleles that exist in CIS (on the same chromosome), representing linked genetic variants. Finally, the Genotype profile extends this concept to encompass collections of related alleles and/or haplotypes that may be in CIS or Trans (on opposite chromosomes) configurations, enabling detailed representation of an individual’s genetic composition. The following is a list of these profiles, their adopted short definitions as well as their corresponding structure definition pages including various examples:
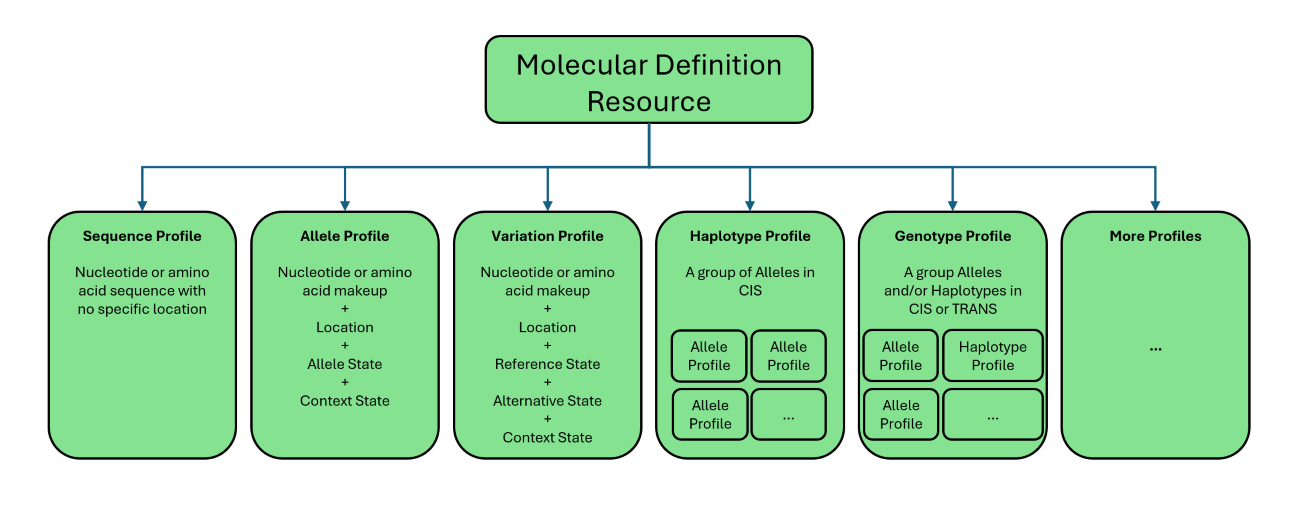
For more details about genomic terms and their corresponding definition, please check this glossary.
This implementation guide is based on the HL7 FHIR standard. It uses terminology, notations and design principles that are specific to FHIR. Before reading this implementation guide, it's important to be familiar with some of the basic principles of FHIR as well as general guidance on how to read FHIR specifications. Readers who are unfamiliar with FHIR are encouraged to read (or at least skim) the following prior to reading the rest of this implementation guide.
Implementers should use this guide to structure genomics data in an interoperable way. The Clinical Genomics Working Group understands that this guide is not complete, and implementers might identify additional concepts and data elements.
| IG | Package | FHIR | Comment |
|---|---|---|---|
| hl7.fhir.uv.molecular-definition-data-types#1.0.0-ballot1 | R6 | ||
| hl7.terminology.r5#7.0.1 | R5 | Automatically added as a dependency - all IGs depend on HL7 Terminology | |
| hl7.fhir.uv.extensions.r5#5.2.0 | R5 | ||
| hl7.fhir.uv.tools.r5#0.9.0 | R5 | for example references |
There are no Global profiles defined
This publication includes IP covered under the following statements.
| Name | Organization |
|---|---|
| Name 1 | Org 1 |
| Name 1 | Org 1 |
| Name 1 | Org 1 |