Molecular Definition Implementation Guide for Molecular Data Types, published by HL7 International / Clinical Genomics. This guide is not an authorized publication; it is the continuous build for version 1.0.0-ballot1 built by the FHIR (HL7® FHIR® Standard) CI Build. This version is based on the current content of https://github.com/HL7/molecular-definition-data-types/ and changes regularly. See the Directory of published versions
| Page standards status: Informative |
Developing a set of specialized MolecularDefinition Profiles to represent discrete genetic data is essential to support clinical use cases that require reliable genetic information across various applications and institutions. The following paragraphs list some of the motivating drivers for this implementation guide.
This implementation guide is not complete. The included artifacts are marked as experimental, but they are ready for review, testing, and validation.
Modern genomic analysis needs cleaner, more expressive data formats than traditional methods provide. Capturing sections of a PDF report is insufficient, as such unstructured data hinders automated processing, integration, and advanced analytics. To fully leverage the power of computational tools and enable scalable, precise interpretation of genetic information, it is imperative to develop a FHIR resource that supports structured, discrete genetic data that convey both content and semantics. This will facilitate more accurate interpretation, seamless data exchange, and robust clinical decision support.
Existing methodologies for representing genomic data within the FHIR framework—encompassing both current resources and profile extensions—fail to sufficiently meet the requirements for discrete genomic data representation. Feedback from implementation specialists indicates that these approaches are characterized by excessive complexity, ambiguity, and inadequate granularity. These deficiencies hinder precise data capture, accurate interpretation, and effective interoperability. Consequently, the inherent complexity impedes the optimal use of genomic data in clinical decision-making and biomedical research contexts. This situation underscores the necessity for a dedicated, optimized FHIR resource designed to enable clear, precise, and streamlined representation of discrete genetic variants, thereby enhancing both implementation feasibility and clinical applicability.
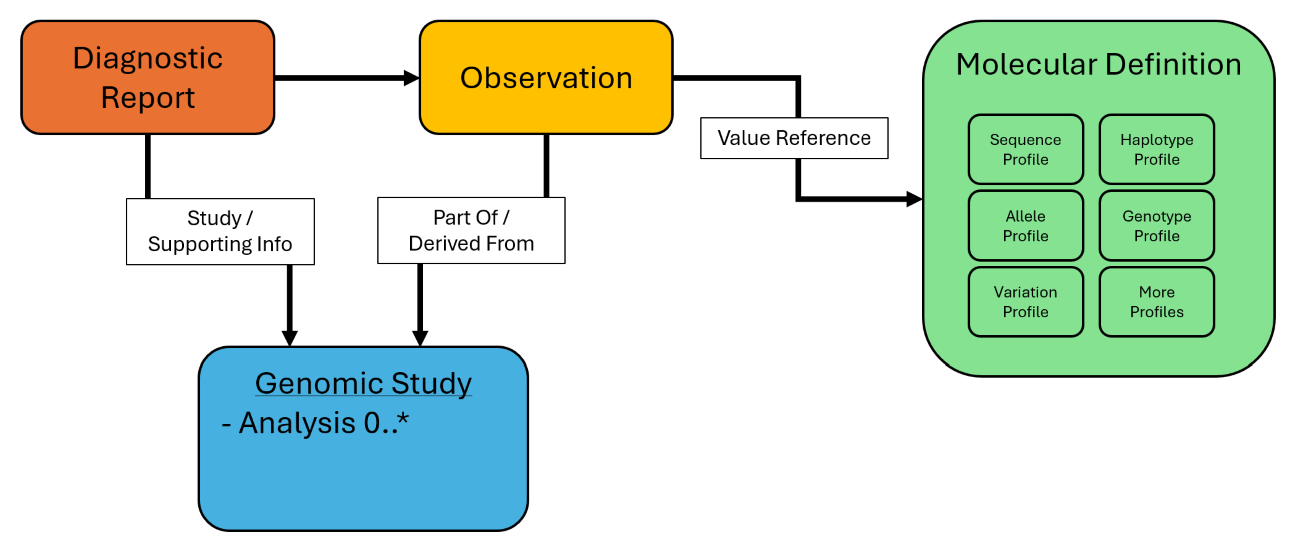
By enabling patient-agnostic FHIR artifacts, MolecularDefinition resource would facilitate linking genetic variants to disease associations, drug interactions, and risks of adverse drug events (ADEs), thereby enhancing precision medicine and pharmacogenomics. In addition, this resource can enable seamless integration of genetic knowledge bases with clinical systems like EHRs, ensuring interoperability and real-time access to up-to-date variant interpretations. Additionally, it could support the reinterpretation of variants as genomic knowledge evolves, allowing clinical decision support systems to provide the most current guidance. This approach addresses current limitations where genetic data is often embedded only within Observations resource instances, restricting broader clinical and research utility.
To ensure broad adoption and interoperability, the MolecularDefinition resource and profiles must support stakeholders beyond HL7, including those involved in national and international genomics initiatives such as ONC’s Sync for Genes and NHGRI’s eMERGE phases 3 and 4. Also, it should align with and facilitate integration with the Global Alliance for Genomics and Health (GA4GH) specifications and driver projects such as:
The MolecularDefinition resource and its specialized profiles offer cleaner data structures and enhanced semantic expression, aligning more naturally with FHIR granular architecture through focused yet connected resources. This design reduces the complexity of existing Observation profiles by reducing dependence on loosely structured Observation components that often obscure the true meaning of genetic data. By enabling attributes to be attached at appropriate levels—such as the report, observation, or variation—the resource preserves semantic clarity and improves data integrity. This focused and well-structured resource not only streamlines implementation but also improves the clarity and utility of genomic information within clinical workflows and interoperable systems.