Genomics Reporting Implementation Guide, published by HL7 International / Clinical Genomics. This guide is not an authorized publication; it is the continuous build for version 4.0.0-ballot built by the FHIR (HL7® FHIR® Standard) CI Build. This version is based on the current content of https://github.com/HL7/genomics-reporting/ and changes regularly. See the Directory of published versions
| Official URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-report | Version: 4.0.0-ballot | |||
| Standards status: Trial-use | Maturity Level: 2 | Computable Name: GenomicReport | ||
Genomic profile of DiagnosticReport.
The genomic report is the focus of all genomic reporting. It conveys metadata about the overall report (what kind of report it was, when it was written, who wrote it, final vs. draft, etc.). It also typically includes a rendered version for review by a clinician. It also groups together all relevant information found as part of the genomic analysis (Rules for relevancy will depend on the type of testing ordered, the reason for testing and the policies of the lab). Most of the structured genomic information is expressed as FHIR Observations. Any recommendations that come with the report are expressed as FHIR Tasks. The report can be organized into sub-reports using core DiagnosticReport extensions like extends or summaryOf, which is especially useful for later analysis steps. Additionally, an observation can be used to group content for viewing purposes or to indicate a higher-level panel (with a specific LOINC panel code in the Observation.code for example).
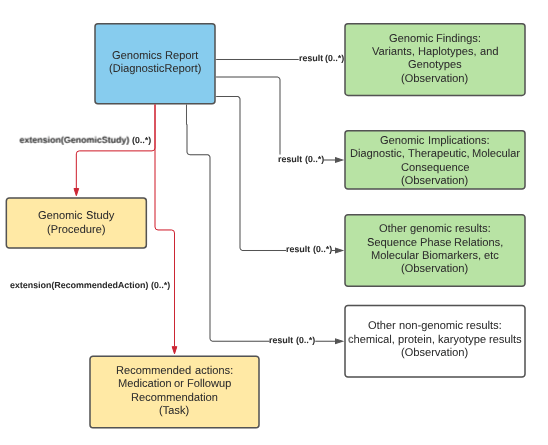
Genomic Report Overview
Usages:
You can also check for usages in the FHIR IG Statistics
Description of Profiles, Differentials, Snapshots and how the different presentations work.
| Name | Flags | Card. | Type | Description & Constraints Filter:   |
|---|---|---|---|---|
 |
0..* | DiagnosticReport | A Diagnostic report - a combination of request information, atomic results, images, interpretation, as well as formatted reports | |
  |
?!Σ | 0..1 | uri | A set of rules under which this content was created |
  |
0..* | Extension | Extension Slice: Unordered, Open by value:url | |
   |
0..* | Reference(Medication Recommendation | Followup Recommendation) | Recommended Action URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/recommended-action | |
   |
0..* | Reference(RiskAssessment) | Genomic Risk Assessment URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-risk-assessment | |
   |
0..* | CodedAnnotation | Comments about the report that also contain a coded type URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-report-note | |
   |
0..* | Reference(Resource) | Other information that may be relevant to this event. URL: http://hl7.org/fhir/StructureDefinition/workflow-supportingInfo | |
   |
0..* | Reference(Genomic Study) | Reference to full details of an genomic study associated with the diagnostic report URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-study-reference | |
   |
0..1 | CodeableConcept | Allele Database URL: http://hl7.org/fhir/StructureDefinition/hla-genotyping-results-allele-database | |
   |
0..1 | (Complex) | glstring URL: http://hl7.org/fhir/StructureDefinition/hla-genotyping-results-glstring | |
   |
0..* | RelatedArtifact | Documentation relevant to the 'parent' resource URL: http://hl7.org/fhir/StructureDefinition/workflow-relatedArtifact | |
  |
?! | 0..* | Extension | Extensions that cannot be ignored |
  |
?!Σ | 1..1 | code | registered | partial | preliminary | final + Binding: DiagnosticReportStatus (required): The status of the diagnostic report. |
  |
Σ | 1..* | CodeableConcept | Service category Slice: Unordered, Open by value:coding Binding: DiagnosticServiceSectionCodes (example): Codes for diagnostic service sections. |
   |
Σ | 1..1 | CodeableConcept | Service category Binding: DiagnosticServiceSectionCodes (example): Codes for diagnostic service sections. |
    |
Σ | 1..1 | Coding | Code defined by a terminology system Required Pattern: At least the following |
     |
0..1 | string | Unique id for inter-element referencing | |
     |
0..* | Extension | Additional content defined by implementations | |
     |
1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/v2-0074 | |
     |
0..1 | string | Version of the system - if relevant | |
     |
1..1 | code | Symbol in syntax defined by the system Fixed Value: GE | |
     |
0..1 | string | Representation defined by the system | |
     |
0..1 | boolean | If this coding was chosen directly by the user | |
  |
Σ | 1..1 | CodeableConcept | Name/Code for this diagnostic report Binding: LOINCDiagnosticReportCodes (preferred): Codes that describe Diagnostic Reports. Required Pattern: At least the following |
   |
0..1 | string | Unique id for inter-element referencing | |
   |
0..* | Extension | Additional content defined by implementations | |
   |
1..* | Coding | Code defined by a terminology system Fixed Value: (Complex) | |
    |
0..1 | string | Unique id for inter-element referencing | |
    |
0..* | Extension | Additional content defined by implementations | |
    |
1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
    |
0..1 | string | Version of the system - if relevant | |
    |
1..1 | code | Symbol in syntax defined by the system Fixed Value: 51969-4 | |
    |
0..1 | string | Representation defined by the system | |
    |
0..1 | boolean | If this coding was chosen directly by the user | |
   |
0..1 | string | Plain text representation of the concept | |
  |
Σ | 0..1 | dateTime | Clinically relevant time/time-period for report |
  |
0..* | Reference(Observation) | Observations Slice: Unordered, Open by profile:resolve() | |
   |
0..* | Reference(Diagnostic Implication) | Diagnostic Implication | |
   |
0..* | Reference(Therapeutic Implication) | Therapeutic Implication | |
   |
0..* | Reference(Molecular Consequence) | Molecular Consequence | |
   |
0..* | Reference(Variant) | Variant | |
   |
0..* | Reference(Sequence Phase Relationship) | Sequence Phase Relationship | |
   |
0..* | Reference(Genotype) | Genotype | |
   |
0..* | Reference(Haplotype) | Haplotype | |
   |
0..* | Reference(Molecular Biomarker) | MolecularBiomarker | |
  |
0..1 | string | Assessment of overall results | |
  |
0..* | CodeableConcept | Coarse overall interpretation of the genomic results Binding: SNOMEDCTClinicalFindings (example): Diagnosis codes provided as adjuncts to the report. | |
 Documentation for this format Documentation for this format | ||||
| Path | Status | Usage | ValueSet | Version | Source |
| DiagnosticReport.status | Base | required | DiagnosticReportStatus | 📍4.0.1 | FHIR Std. |
| DiagnosticReport.category | Base | example | Diagnostic Service Section Codes | 📍4.0.1 | FHIR Std. |
| DiagnosticReport.category:Genetics | Base | example | Diagnostic Service Section Codes | 📍4.0.1 | FHIR Std. |
| DiagnosticReport.code | Base | preferred | LOINC Diagnostic Report Codes | 📍4.0.1 | FHIR Std. |
| DiagnosticReport.conclusionCode | Base | example | SNOMED CT Clinical Findings | 📍4.0.1 | FHIR Std. |
| Id | Grade | Path(s) | Description | Expression |
| dom-2 | error | DiagnosticReport | If the resource is contained in another resource, it SHALL NOT contain nested Resources |
contained.contained.empty()
|
| dom-3 | error | DiagnosticReport | If the resource is contained in another resource, it SHALL be referred to from elsewhere in the resource or SHALL refer to the containing resource |
contained.where((('#'+id in (%resource.descendants().reference | %resource.descendants().as(canonical) | %resource.descendants().as(uri) | %resource.descendants().as(url))) or descendants().where(reference = '#').exists() or descendants().where(as(canonical) = '#').exists() or descendants().where(as(canonical) = '#').exists()).not()).trace('unmatched', id).empty()
|
| dom-4 | error | DiagnosticReport | If a resource is contained in another resource, it SHALL NOT have a meta.versionId or a meta.lastUpdated |
contained.meta.versionId.empty() and contained.meta.lastUpdated.empty()
|
| dom-5 | error | DiagnosticReport | If a resource is contained in another resource, it SHALL NOT have a security label |
contained.meta.security.empty()
|
| dom-6 | best practice | DiagnosticReport | A resource should have narrative for robust management |
text.`div`.exists()
|
| ele-1 | error | **ALL** elements | All FHIR elements must have a @value or children |
hasValue() or (children().count() > id.count())
|
| ext-1 | error | **ALL** extensions | Must have either extensions or value[x], not both |
extension.exists() != value.exists()
|
This structure is derived from DiagnosticReport
| Name | Flags | Card. | Type | Description & Constraints Filter:   | ||||
|---|---|---|---|---|---|---|---|---|
 |
0..* | DiagnosticReport | A Diagnostic report - a combination of request information, atomic results, images, interpretation, as well as formatted reports | |||||
  |
Σ | 0..1 | id | Logical id of this artifact | ||||
  |
Σ | 0..1 | Meta | Metadata about the resource | ||||
  |
?!Σ | 0..1 | uri | A set of rules under which this content was created | ||||
  |
0..1 | code | Language of the resource content Binding: CommonLanguages (preferred): A human language.
| |||||
  |
0..1 | Narrative | Text summary of the resource, for human interpretation This profile does not constrain the narrative in regard to content, language, or traceability to data elements | |||||
  |
0..* | Resource | Contained, inline Resources | |||||
  |
0..* | Extension | Extension Slice: Unordered, Open by value:url | |||||
   |
0..* | Reference(Medication Recommendation | Followup Recommendation) | Recommended Action URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/recommended-action | |||||
   |
0..* | Reference(RiskAssessment) | Genomic Risk Assessment URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-risk-assessment | |||||
   |
0..* | CodedAnnotation | Comments about the report that also contain a coded type URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-report-note | |||||
   |
0..* | Reference(Resource) | Other information that may be relevant to this event. URL: http://hl7.org/fhir/StructureDefinition/workflow-supportingInfo | |||||
   |
0..* | Reference(Genomic Study) | Reference to full details of an genomic study associated with the diagnostic report URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-study-reference | |||||
   |
0..1 | CodeableConcept | Allele Database URL: http://hl7.org/fhir/StructureDefinition/hla-genotyping-results-allele-database | |||||
   |
0..1 | (Complex) | glstring URL: http://hl7.org/fhir/StructureDefinition/hla-genotyping-results-glstring | |||||
   |
0..* | RelatedArtifact | Documentation relevant to the 'parent' resource URL: http://hl7.org/fhir/StructureDefinition/workflow-relatedArtifact | |||||
  |
?! | 0..* | Extension | Extensions that cannot be ignored | ||||
  |
Σ | 0..* | Identifier | Business identifier for report | ||||
  |
0..* | Reference(CarePlan | ImmunizationRecommendation | MedicationRequest | NutritionOrder | ServiceRequest) | What was requested | |||||
  |
?!Σ | 1..1 | code | registered | partial | preliminary | final + Binding: DiagnosticReportStatus (required): The status of the diagnostic report. | ||||
  |
Σ | 1..* | CodeableConcept | Service category Slice: Unordered, Open by value:coding Binding: DiagnosticServiceSectionCodes (example): Codes for diagnostic service sections. | ||||
   |
Σ | 1..1 | CodeableConcept | Service category Binding: DiagnosticServiceSectionCodes (example): Codes for diagnostic service sections. | ||||
    |
0..1 | string | Unique id for inter-element referencing | |||||
    |
0..* | Extension | Additional content defined by implementations Slice: Unordered, Open by value:url | |||||
    |
Σ | 1..1 | Coding | Code defined by a terminology system Required Pattern: At least the following | ||||
     |
0..1 | string | Unique id for inter-element referencing | |||||
     |
0..* | Extension | Additional content defined by implementations | |||||
     |
1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/v2-0074 | |||||
     |
0..1 | string | Version of the system - if relevant | |||||
     |
1..1 | code | Symbol in syntax defined by the system Fixed Value: GE | |||||
     |
0..1 | string | Representation defined by the system | |||||
     |
0..1 | boolean | If this coding was chosen directly by the user | |||||
    |
Σ | 0..1 | string | Plain text representation of the concept | ||||
  |
Σ | 1..1 | CodeableConcept | Name/Code for this diagnostic report Binding: LOINCDiagnosticReportCodes (preferred): Codes that describe Diagnostic Reports. Required Pattern: At least the following | ||||
   |
0..1 | string | Unique id for inter-element referencing | |||||
   |
0..* | Extension | Additional content defined by implementations | |||||
   |
1..* | Coding | Code defined by a terminology system Fixed Value: (Complex) | |||||
    |
0..1 | string | Unique id for inter-element referencing | |||||
    |
0..* | Extension | Additional content defined by implementations | |||||
    |
1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
    |
0..1 | string | Version of the system - if relevant | |||||
    |
1..1 | code | Symbol in syntax defined by the system Fixed Value: 51969-4 | |||||
    |
0..1 | string | Representation defined by the system | |||||
    |
0..1 | boolean | If this coding was chosen directly by the user | |||||
   |
0..1 | string | Plain text representation of the concept | |||||
  |
Σ | 0..1 | Reference(Patient | Group | Device | Location) | The subject of the report - usually, but not always, the patient | ||||
  |
Σ | 0..1 | Reference(Encounter) | Health care event when test ordered | ||||
  |
Σ | 0..1 | dateTime | Clinically relevant time/time-period for report | ||||
  |
Σ | 0..1 | instant | DateTime this version was made | ||||
  |
Σ | 0..* | Reference(Practitioner | PractitionerRole | Organization | CareTeam) | Responsible Diagnostic Service | ||||
  |
Σ | 0..* | Reference(Practitioner | PractitionerRole | Organization | CareTeam) | Primary result interpreter | ||||
  |
0..* | Reference(Specimen) | Specimens this report is based on | |||||
  |
0..* | Reference(Observation) | Observations Slice: Unordered, Open by profile:resolve() | |||||
   |
0..* | Reference(Diagnostic Implication) | Diagnostic Implication | |||||
   |
0..* | Reference(Therapeutic Implication) | Therapeutic Implication | |||||
   |
0..* | Reference(Molecular Consequence) | Molecular Consequence | |||||
   |
0..* | Reference(Variant) | Variant | |||||
   |
0..* | Reference(Sequence Phase Relationship) | Sequence Phase Relationship | |||||
   |
0..* | Reference(Genotype) | Genotype | |||||
   |
0..* | Reference(Haplotype) | Haplotype | |||||
   |
0..* | Reference(Molecular Biomarker) | MolecularBiomarker | |||||
  |
0..* | Reference(ImagingStudy) | Reference to full details of imaging associated with the diagnostic report | |||||
  |
Σ | 0..* | BackboneElement | Key images associated with this report | ||||
   |
0..1 | string | Unique id for inter-element referencing | |||||
   |
0..* | Extension | Additional content defined by implementations | |||||
   |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
   |
0..1 | string | Comment about the image (e.g. explanation) | |||||
   |
Σ | 1..1 | Reference(Media) | Reference to the image source | ||||
  |
0..1 | string | Assessment of overall results | |||||
  |
0..* | CodeableConcept | Coarse overall interpretation of the genomic results Binding: SNOMEDCTClinicalFindings (example): Diagnosis codes provided as adjuncts to the report. | |||||
  |
0..* | Attachment | Entire report as issued | |||||
 Documentation for this format Documentation for this format | ||||||||
| Path | Status | Usage | ValueSet | Version | Source |
| DiagnosticReport.language | Base | preferred | Common Languages | 📍4.0.1 | FHIR Std. |
| DiagnosticReport.status | Base | required | DiagnosticReportStatus | 📍4.0.1 | FHIR Std. |
| DiagnosticReport.category | Base | example | Diagnostic Service Section Codes | 📍4.0.1 | FHIR Std. |
| DiagnosticReport.category:Genetics | Base | example | Diagnostic Service Section Codes | 📍4.0.1 | FHIR Std. |
| DiagnosticReport.code | Base | preferred | LOINC Diagnostic Report Codes | 📍4.0.1 | FHIR Std. |
| DiagnosticReport.conclusionCode | Base | example | SNOMED CT Clinical Findings | 📍4.0.1 | FHIR Std. |
| Id | Grade | Path(s) | Description | Expression |
| dom-2 | error | DiagnosticReport | If the resource is contained in another resource, it SHALL NOT contain nested Resources |
contained.contained.empty()
|
| dom-3 | error | DiagnosticReport | If the resource is contained in another resource, it SHALL be referred to from elsewhere in the resource or SHALL refer to the containing resource |
contained.where((('#'+id in (%resource.descendants().reference | %resource.descendants().as(canonical) | %resource.descendants().as(uri) | %resource.descendants().as(url))) or descendants().where(reference = '#').exists() or descendants().where(as(canonical) = '#').exists() or descendants().where(as(canonical) = '#').exists()).not()).trace('unmatched', id).empty()
|
| dom-4 | error | DiagnosticReport | If a resource is contained in another resource, it SHALL NOT have a meta.versionId or a meta.lastUpdated |
contained.meta.versionId.empty() and contained.meta.lastUpdated.empty()
|
| dom-5 | error | DiagnosticReport | If a resource is contained in another resource, it SHALL NOT have a security label |
contained.meta.security.empty()
|
| dom-6 | best practice | DiagnosticReport | A resource should have narrative for robust management |
text.`div`.exists()
|
| ele-1 | error | **ALL** elements | All FHIR elements must have a @value or children |
hasValue() or (children().count() > id.count())
|
| ext-1 | error | **ALL** extensions | Must have either extensions or value[x], not both |
extension.exists() != value.exists()
|
This structure is derived from DiagnosticReport
Summary
Mandatory: 3 elements
Structures
This structure refers to these other structures:
Extensions
This structure refers to these extensions:
Slices
This structure defines the following Slices:
Maturity: 2
Key Elements View
| Name | Flags | Card. | Type | Description & Constraints Filter:   |
|---|---|---|---|---|
 |
0..* | DiagnosticReport | A Diagnostic report - a combination of request information, atomic results, images, interpretation, as well as formatted reports | |
  |
?!Σ | 0..1 | uri | A set of rules under which this content was created |
  |
0..* | Extension | Extension Slice: Unordered, Open by value:url | |
   |
0..* | Reference(Medication Recommendation | Followup Recommendation) | Recommended Action URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/recommended-action | |
   |
0..* | Reference(RiskAssessment) | Genomic Risk Assessment URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-risk-assessment | |
   |
0..* | CodedAnnotation | Comments about the report that also contain a coded type URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-report-note | |
   |
0..* | Reference(Resource) | Other information that may be relevant to this event. URL: http://hl7.org/fhir/StructureDefinition/workflow-supportingInfo | |
   |
0..* | Reference(Genomic Study) | Reference to full details of an genomic study associated with the diagnostic report URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-study-reference | |
   |
0..1 | CodeableConcept | Allele Database URL: http://hl7.org/fhir/StructureDefinition/hla-genotyping-results-allele-database | |
   |
0..1 | (Complex) | glstring URL: http://hl7.org/fhir/StructureDefinition/hla-genotyping-results-glstring | |
   |
0..* | RelatedArtifact | Documentation relevant to the 'parent' resource URL: http://hl7.org/fhir/StructureDefinition/workflow-relatedArtifact | |
  |
?! | 0..* | Extension | Extensions that cannot be ignored |
  |
?!Σ | 1..1 | code | registered | partial | preliminary | final + Binding: DiagnosticReportStatus (required): The status of the diagnostic report. |
  |
Σ | 1..* | CodeableConcept | Service category Slice: Unordered, Open by value:coding Binding: DiagnosticServiceSectionCodes (example): Codes for diagnostic service sections. |
   |
Σ | 1..1 | CodeableConcept | Service category Binding: DiagnosticServiceSectionCodes (example): Codes for diagnostic service sections. |
    |
Σ | 1..1 | Coding | Code defined by a terminology system Required Pattern: At least the following |
     |
0..1 | string | Unique id for inter-element referencing | |
     |
0..* | Extension | Additional content defined by implementations | |
     |
1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/v2-0074 | |
     |
0..1 | string | Version of the system - if relevant | |
     |
1..1 | code | Symbol in syntax defined by the system Fixed Value: GE | |
     |
0..1 | string | Representation defined by the system | |
     |
0..1 | boolean | If this coding was chosen directly by the user | |
  |
Σ | 1..1 | CodeableConcept | Name/Code for this diagnostic report Binding: LOINCDiagnosticReportCodes (preferred): Codes that describe Diagnostic Reports. Required Pattern: At least the following |
   |
0..1 | string | Unique id for inter-element referencing | |
   |
0..* | Extension | Additional content defined by implementations | |
   |
1..* | Coding | Code defined by a terminology system Fixed Value: (Complex) | |
    |
0..1 | string | Unique id for inter-element referencing | |
    |
0..* | Extension | Additional content defined by implementations | |
    |
1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
    |
0..1 | string | Version of the system - if relevant | |
    |
1..1 | code | Symbol in syntax defined by the system Fixed Value: 51969-4 | |
    |
0..1 | string | Representation defined by the system | |
    |
0..1 | boolean | If this coding was chosen directly by the user | |
   |
0..1 | string | Plain text representation of the concept | |
  |
Σ | 0..1 | dateTime | Clinically relevant time/time-period for report |
  |
0..* | Reference(Observation) | Observations Slice: Unordered, Open by profile:resolve() | |
   |
0..* | Reference(Diagnostic Implication) | Diagnostic Implication | |
   |
0..* | Reference(Therapeutic Implication) | Therapeutic Implication | |
   |
0..* | Reference(Molecular Consequence) | Molecular Consequence | |
   |
0..* | Reference(Variant) | Variant | |
   |
0..* | Reference(Sequence Phase Relationship) | Sequence Phase Relationship | |
   |
0..* | Reference(Genotype) | Genotype | |
   |
0..* | Reference(Haplotype) | Haplotype | |
   |
0..* | Reference(Molecular Biomarker) | MolecularBiomarker | |
  |
0..1 | string | Assessment of overall results | |
  |
0..* | CodeableConcept | Coarse overall interpretation of the genomic results Binding: SNOMEDCTClinicalFindings (example): Diagnosis codes provided as adjuncts to the report. | |
 Documentation for this format Documentation for this format | ||||
| Path | Status | Usage | ValueSet | Version | Source |
| DiagnosticReport.status | Base | required | DiagnosticReportStatus | 📍4.0.1 | FHIR Std. |
| DiagnosticReport.category | Base | example | Diagnostic Service Section Codes | 📍4.0.1 | FHIR Std. |
| DiagnosticReport.category:Genetics | Base | example | Diagnostic Service Section Codes | 📍4.0.1 | FHIR Std. |
| DiagnosticReport.code | Base | preferred | LOINC Diagnostic Report Codes | 📍4.0.1 | FHIR Std. |
| DiagnosticReport.conclusionCode | Base | example | SNOMED CT Clinical Findings | 📍4.0.1 | FHIR Std. |
| Id | Grade | Path(s) | Description | Expression |
| dom-2 | error | DiagnosticReport | If the resource is contained in another resource, it SHALL NOT contain nested Resources |
contained.contained.empty()
|
| dom-3 | error | DiagnosticReport | If the resource is contained in another resource, it SHALL be referred to from elsewhere in the resource or SHALL refer to the containing resource |
contained.where((('#'+id in (%resource.descendants().reference | %resource.descendants().as(canonical) | %resource.descendants().as(uri) | %resource.descendants().as(url))) or descendants().where(reference = '#').exists() or descendants().where(as(canonical) = '#').exists() or descendants().where(as(canonical) = '#').exists()).not()).trace('unmatched', id).empty()
|
| dom-4 | error | DiagnosticReport | If a resource is contained in another resource, it SHALL NOT have a meta.versionId or a meta.lastUpdated |
contained.meta.versionId.empty() and contained.meta.lastUpdated.empty()
|
| dom-5 | error | DiagnosticReport | If a resource is contained in another resource, it SHALL NOT have a security label |
contained.meta.security.empty()
|
| dom-6 | best practice | DiagnosticReport | A resource should have narrative for robust management |
text.`div`.exists()
|
| ele-1 | error | **ALL** elements | All FHIR elements must have a @value or children |
hasValue() or (children().count() > id.count())
|
| ext-1 | error | **ALL** extensions | Must have either extensions or value[x], not both |
extension.exists() != value.exists()
|
Differential View
This structure is derived from DiagnosticReport
Snapshot View
| Name | Flags | Card. | Type | Description & Constraints Filter:   | ||||
|---|---|---|---|---|---|---|---|---|
 |
0..* | DiagnosticReport | A Diagnostic report - a combination of request information, atomic results, images, interpretation, as well as formatted reports | |||||
  |
Σ | 0..1 | id | Logical id of this artifact | ||||
  |
Σ | 0..1 | Meta | Metadata about the resource | ||||
  |
?!Σ | 0..1 | uri | A set of rules under which this content was created | ||||
  |
0..1 | code | Language of the resource content Binding: CommonLanguages (preferred): A human language.
| |||||
  |
0..1 | Narrative | Text summary of the resource, for human interpretation This profile does not constrain the narrative in regard to content, language, or traceability to data elements | |||||
  |
0..* | Resource | Contained, inline Resources | |||||
  |
0..* | Extension | Extension Slice: Unordered, Open by value:url | |||||
   |
0..* | Reference(Medication Recommendation | Followup Recommendation) | Recommended Action URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/recommended-action | |||||
   |
0..* | Reference(RiskAssessment) | Genomic Risk Assessment URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-risk-assessment | |||||
   |
0..* | CodedAnnotation | Comments about the report that also contain a coded type URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-report-note | |||||
   |
0..* | Reference(Resource) | Other information that may be relevant to this event. URL: http://hl7.org/fhir/StructureDefinition/workflow-supportingInfo | |||||
   |
0..* | Reference(Genomic Study) | Reference to full details of an genomic study associated with the diagnostic report URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-study-reference | |||||
   |
0..1 | CodeableConcept | Allele Database URL: http://hl7.org/fhir/StructureDefinition/hla-genotyping-results-allele-database | |||||
   |
0..1 | (Complex) | glstring URL: http://hl7.org/fhir/StructureDefinition/hla-genotyping-results-glstring | |||||
   |
0..* | RelatedArtifact | Documentation relevant to the 'parent' resource URL: http://hl7.org/fhir/StructureDefinition/workflow-relatedArtifact | |||||
  |
?! | 0..* | Extension | Extensions that cannot be ignored | ||||
  |
Σ | 0..* | Identifier | Business identifier for report | ||||
  |
0..* | Reference(CarePlan | ImmunizationRecommendation | MedicationRequest | NutritionOrder | ServiceRequest) | What was requested | |||||
  |
?!Σ | 1..1 | code | registered | partial | preliminary | final + Binding: DiagnosticReportStatus (required): The status of the diagnostic report. | ||||
  |
Σ | 1..* | CodeableConcept | Service category Slice: Unordered, Open by value:coding Binding: DiagnosticServiceSectionCodes (example): Codes for diagnostic service sections. | ||||
   |
Σ | 1..1 | CodeableConcept | Service category Binding: DiagnosticServiceSectionCodes (example): Codes for diagnostic service sections. | ||||
    |
0..1 | string | Unique id for inter-element referencing | |||||
    |
0..* | Extension | Additional content defined by implementations Slice: Unordered, Open by value:url | |||||
    |
Σ | 1..1 | Coding | Code defined by a terminology system Required Pattern: At least the following | ||||
     |
0..1 | string | Unique id for inter-element referencing | |||||
     |
0..* | Extension | Additional content defined by implementations | |||||
     |
1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/v2-0074 | |||||
     |
0..1 | string | Version of the system - if relevant | |||||
     |
1..1 | code | Symbol in syntax defined by the system Fixed Value: GE | |||||
     |
0..1 | string | Representation defined by the system | |||||
     |
0..1 | boolean | If this coding was chosen directly by the user | |||||
    |
Σ | 0..1 | string | Plain text representation of the concept | ||||
  |
Σ | 1..1 | CodeableConcept | Name/Code for this diagnostic report Binding: LOINCDiagnosticReportCodes (preferred): Codes that describe Diagnostic Reports. Required Pattern: At least the following | ||||
   |
0..1 | string | Unique id for inter-element referencing | |||||
   |
0..* | Extension | Additional content defined by implementations | |||||
   |
1..* | Coding | Code defined by a terminology system Fixed Value: (Complex) | |||||
    |
0..1 | string | Unique id for inter-element referencing | |||||
    |
0..* | Extension | Additional content defined by implementations | |||||
    |
1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
    |
0..1 | string | Version of the system - if relevant | |||||
    |
1..1 | code | Symbol in syntax defined by the system Fixed Value: 51969-4 | |||||
    |
0..1 | string | Representation defined by the system | |||||
    |
0..1 | boolean | If this coding was chosen directly by the user | |||||
   |
0..1 | string | Plain text representation of the concept | |||||
  |
Σ | 0..1 | Reference(Patient | Group | Device | Location) | The subject of the report - usually, but not always, the patient | ||||
  |
Σ | 0..1 | Reference(Encounter) | Health care event when test ordered | ||||
  |
Σ | 0..1 | dateTime | Clinically relevant time/time-period for report | ||||
  |
Σ | 0..1 | instant | DateTime this version was made | ||||
  |
Σ | 0..* | Reference(Practitioner | PractitionerRole | Organization | CareTeam) | Responsible Diagnostic Service | ||||
  |
Σ | 0..* | Reference(Practitioner | PractitionerRole | Organization | CareTeam) | Primary result interpreter | ||||
  |
0..* | Reference(Specimen) | Specimens this report is based on | |||||
  |
0..* | Reference(Observation) | Observations Slice: Unordered, Open by profile:resolve() | |||||
   |
0..* | Reference(Diagnostic Implication) | Diagnostic Implication | |||||
   |
0..* | Reference(Therapeutic Implication) | Therapeutic Implication | |||||
   |
0..* | Reference(Molecular Consequence) | Molecular Consequence | |||||
   |
0..* | Reference(Variant) | Variant | |||||
   |
0..* | Reference(Sequence Phase Relationship) | Sequence Phase Relationship | |||||
   |
0..* | Reference(Genotype) | Genotype | |||||
   |
0..* | Reference(Haplotype) | Haplotype | |||||
   |
0..* | Reference(Molecular Biomarker) | MolecularBiomarker | |||||
  |
0..* | Reference(ImagingStudy) | Reference to full details of imaging associated with the diagnostic report | |||||
  |
Σ | 0..* | BackboneElement | Key images associated with this report | ||||
   |
0..1 | string | Unique id for inter-element referencing | |||||
   |
0..* | Extension | Additional content defined by implementations | |||||
   |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
   |
0..1 | string | Comment about the image (e.g. explanation) | |||||
   |
Σ | 1..1 | Reference(Media) | Reference to the image source | ||||
  |
0..1 | string | Assessment of overall results | |||||
  |
0..* | CodeableConcept | Coarse overall interpretation of the genomic results Binding: SNOMEDCTClinicalFindings (example): Diagnosis codes provided as adjuncts to the report. | |||||
  |
0..* | Attachment | Entire report as issued | |||||
 Documentation for this format Documentation for this format | ||||||||
| Path | Status | Usage | ValueSet | Version | Source |
| DiagnosticReport.language | Base | preferred | Common Languages | 📍4.0.1 | FHIR Std. |
| DiagnosticReport.status | Base | required | DiagnosticReportStatus | 📍4.0.1 | FHIR Std. |
| DiagnosticReport.category | Base | example | Diagnostic Service Section Codes | 📍4.0.1 | FHIR Std. |
| DiagnosticReport.category:Genetics | Base | example | Diagnostic Service Section Codes | 📍4.0.1 | FHIR Std. |
| DiagnosticReport.code | Base | preferred | LOINC Diagnostic Report Codes | 📍4.0.1 | FHIR Std. |
| DiagnosticReport.conclusionCode | Base | example | SNOMED CT Clinical Findings | 📍4.0.1 | FHIR Std. |
| Id | Grade | Path(s) | Description | Expression |
| dom-2 | error | DiagnosticReport | If the resource is contained in another resource, it SHALL NOT contain nested Resources |
contained.contained.empty()
|
| dom-3 | error | DiagnosticReport | If the resource is contained in another resource, it SHALL be referred to from elsewhere in the resource or SHALL refer to the containing resource |
contained.where((('#'+id in (%resource.descendants().reference | %resource.descendants().as(canonical) | %resource.descendants().as(uri) | %resource.descendants().as(url))) or descendants().where(reference = '#').exists() or descendants().where(as(canonical) = '#').exists() or descendants().where(as(canonical) = '#').exists()).not()).trace('unmatched', id).empty()
|
| dom-4 | error | DiagnosticReport | If a resource is contained in another resource, it SHALL NOT have a meta.versionId or a meta.lastUpdated |
contained.meta.versionId.empty() and contained.meta.lastUpdated.empty()
|
| dom-5 | error | DiagnosticReport | If a resource is contained in another resource, it SHALL NOT have a security label |
contained.meta.security.empty()
|
| dom-6 | best practice | DiagnosticReport | A resource should have narrative for robust management |
text.`div`.exists()
|
| ele-1 | error | **ALL** elements | All FHIR elements must have a @value or children |
hasValue() or (children().count() > id.count())
|
| ext-1 | error | **ALL** extensions | Must have either extensions or value[x], not both |
extension.exists() != value.exists()
|
This structure is derived from DiagnosticReport
Summary
Mandatory: 3 elements
Structures
This structure refers to these other structures:
Extensions
This structure refers to these extensions:
Slices
This structure defines the following Slices:
Maturity: 2
Other representations of profile: CSV, Excel, Schematron
On the report, it is important to answer the question "Did you find anything when you did the test I asked you to do?" The data sender can use attributes of conclusionCode and conclusion to represent the summary result of the test (e.g., Positive, Negative, Unknown) and a textual summary. These are typically used when the genomic test was looking for a particular genomically-based disease. It allows indication of whether genomic results known to be associated with the disease were found or not.
Results observation profiles, like genomic observations, are typically referenced directly by a Genomic Report. The genetic findings and implication profiles all contain links to computably define their composite relationships (e.g., the variant observation is referenced within the implication profile using derivedFrom). However, observations could be organized into groups by other observations. See this grouping guidance for an overview with examples and considerations for processing reports. Be aware that consumers of Genomic Diagnostic Report MUST navigate through all hasMember relations and navigate through derivedFrom relationships to ensure processing of all clinically relevant information.
In some cases, the lab or other reporting organization may generate risk assessments as part of their reports. These are referenced from a report or an observation from the Genomic Risk extension.
When sending a copy of the report (e.g., PDF or other document containing the written report), use presentedForm. Note this is different from the Related Artifact extension, which is used to reference citations, evidence and other supporting documentation for the observation or report. Another approach which should be avoided (at least for this current release) is the DiagnosticReport.media attribute. Its definition focuses on "Key images associated with this report" which does not align well with this use case.
If needed, large or complex genomic reports may be broken down into sub-reports using core DiagnosticReport extensions like extends or summaryOf. This approach is particularly useful when different labs or services are performing later steps in the analysis, for example. Or a panel Observation.code can be used.