NHS North West Genomics
0.1.0 - ci-build
NHS North West Genomics
0.1.0 - ci-build
NHS North West Genomics, published by NHS North West Genomics. This guide is not an authorized publication; it is the continuous build for version 0.1.0 built by the FHIR (HL7® FHIR® Standard) CI Build. This version is based on the current content of https://github.com/nw-gmsa/nw-gmsa.github.com/ and changes regularly. See the Directory of published versions
This guide is to support Genomic Testing Workflow at a regional level and is designed to be compatible with:
The general workflow is based on IHE LTW profiles and HL7 v2 OML and ORU.
Genomic Testing Workflow is part of Diagnostic Testing, which is also part of the general clinical process.
graph TD;
A[Assessment]-->|Creates Observations| B;
A--> |Needs Diagnostic Testing and Completes| T;
B[Diagnosis]-->|Creates Condition| C;
T[<b>Order Placer</b><br/>Genomics Test Order]--> |"Sends Laboratory Order<br/>LAB-1 FHIR Message O21"| AN;
T --> |Asks for| S
S[Specimen Collection] --> |Sends Specimen| AN;
AN["<b>Order Filler</b><br/>Diagnostic Testing"] --> |"Requests further tests <br/>(reflex order)"| T;
AN --> |Sends Laboratory Report<br/>LAB-3 HL7 v2 ORU_R01| A;
C[Plan]-->|Creates Goals and Tasks| D;
D[Implement/Interventions]-->|Actions Tasks| E;
E[Evaluate]--> |Reviews Care| A;
click T Questionnaire-GenomicTestOrder.html
click AN Questionnaire-GenomicTestReport.html
click S ExampleScenario-BiopsyProcedure.html
classDef purple fill:#E1D5E7;
classDef yellow fill:#FFF2CC;
classDef pink fill:#F8CECC
classDef green fill:#D5E8D4;
classDef blue fill:#DAE8FC;
classDef orange fill:#FFE6CC;
class A pink
class B yellow
class C green
class D blue
class E orange
class O,S,T,AN purple
Genomic diagnostic testing follows the same standardized process defined by the IHE Laboratory Testing Workflow used in traditional laboratory testing. This workflow has been enhanced to support the sharing of laboratory reports (documents) through Integrated Care Systems (ICS). In addition, a new mechanism for sharing laboratory reports has been introduced to establish a regional genomic data repository.
graph TD;
subgraph NHSTrust[NHS Trust]
T[<b>Order Placer</b><br/>EPR]--> |"1a. Sends Laboratory Order<br>LAB-1 HL7v2 ORM_O01/OML_O21"| TIE;
TIE[Trust Integration Engine]
TIE--> |4c. Sends Laboratory Report<br/>LAB-3 HL7 v2 ORU_R01| T;
end
TIE --> |"1b. Sends Laboratory Order<br>LAB-1 FHIR Message O21"| RIE;
T --> |2. Asks for| S
S[Specimen Collection] --> |3. Sends Specimen| AN;
subgraph NWGenomics[North West Genomics]
RIE --> |"1c. Sends Laboratory Order<br>LAB-1 HL7 v2 OML_O21"| AN;
AN["<b>Order Filler</b><br/>Diagnostic Testing<br/>LIMS iGene"] --> |4a. Sends Laboratory Report<br/>LAB-3 HL7 v2 ORU_R01| RIE;
RIE[Regional Integration Engine] --> |4b. Sends Laboratory Report<br/>LAB-3 HL7 v2 ORU_R01| TIE;
end
click T Questionnaire-GenomicTestOrder.html
click AN Questionnaire-GenomicTestReport.html
click S ExampleScenario-BiopsyProcedure.html
classDef purple fill:#E1D5E7;
classDef yellow fill:#FFF2CC;
classDef pink fill:#F8CECC
classDef green fill:#D5E8D4;
classDef blue fill:#DAE8FC;
classDef orange fill:#FFE6CC;
class A pink
class B yellow
class C green
class D blue
class E orange
class O,S,T,AN purple
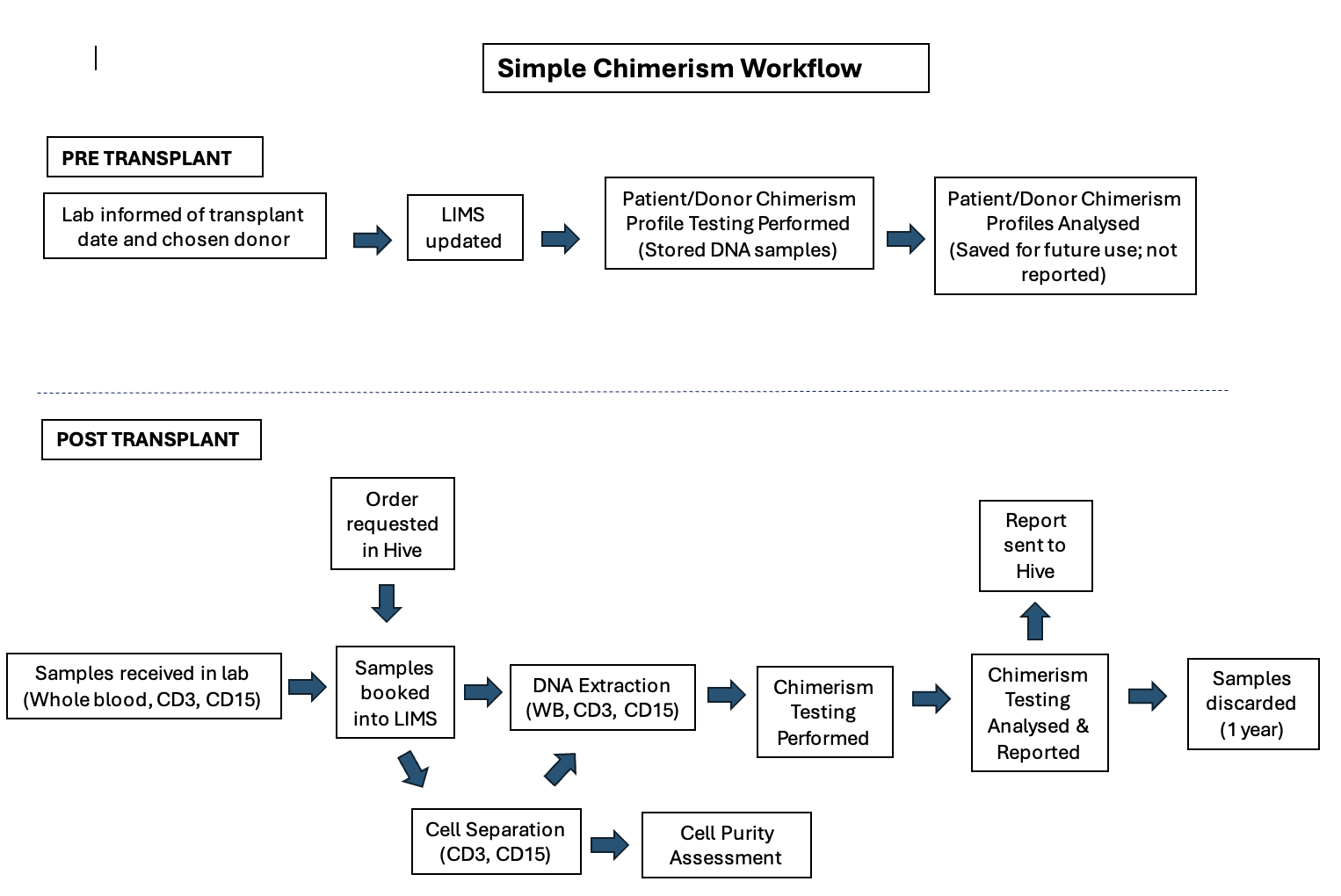
The chimerism testing pathway within the NHS is used to monitor the success of a haematopoietic stem cell transplant (HSCT) and involves a specific process of sample collection, transport, and analysis guided by clinical consensus guidelines. The testing is requested by a patient's clinical team, not directly by the patient.
graph TD;
subgraph NHSTrust[NHS Trust]
Practitioner[Consultant Haematologist<br/>Practitioner] --> |1. Creates Order| EPR[<b>Order Placer</b><br/>EPR]
Practitioner --> |3. Asks For| Specimen[Sample Collection]
TIE[Trust Integration Engine]
end
EPR --> |"2a. Sends Laboratory Order<br/>M118.1 Short Tandem Repeat (STR)<br/>HL7 v2 ORM_O01 (MFT)"| TIE
Specimen --> |4. Send Specimen| LIMS
TIE --> |"2b. Sends Laboratory Order<br/>HL7 v2 ORM_O01"| RIE
subgraph NWGenomics[North West Genomics]
RIE --> |"2b. Sends Laboratory Order<br/>HL7 v2 ORM_O01"| LIMS
LIMS[<b>Order Filler</b><br/>LIMS Histotrac]
LIMS --> |5a. Send Laboratory Report<br/>HL7 v2 ORU_R01| RIE["Regional Integration Engine"]
end
RIE --> |5b. Send Laboratory Report<br/>HL7 v2 ORU_R01| TIE
TIE --> |5c. Send Laboratory Report<br/>HL7 v2 ORU_R01| EPR
classDef purple fill:#E1D5E7;
class EPR,LIMS,Specimen,Practitioner purple
Chimerism Genomic Tests - MFT
Future
graph TD
subgraph Trust[NHS Trust]
EPR[<b>Order Placer</b><br/>EPR]
TIE[Trust Integration Engine]
end
EPR --> |"1. Create Laboratory Order<br/>Manual entry"| HODS
HODS --> |"2. Send Laboratory Order + Specimen<br/>"| MFTReception[Specimen Reception]
MFTReception --> |"3a. (Manual) Immunology Laboratory Order + Specimen"| LIMS["<b>Order Filler</b><br/>Immunology LIMS"]
subgraph Laboratory["Laboratory - MFT"]
LIMS --> |3b. Send Laboratory Report<br/>HL7 v2 ORU_R01| LIE[Laboratory<br/>Trust Integration Engine]
end
LIE --> |3c. Send Laboratory Report<br/>HL7 v2 ORU_R01| RIE
RIE --> |3d and 4d. Send Laboratory Report<br/>HL7 v2 ORU_R01| HODS
MFTReception --> |"4a. Genomics Laboratory Order + Specimen<br/>Manual"| TestType
subgraph NWGenomics[North West Genomics]
RIE["Regional Integration Engine"]
HODS["<b>Order Filler</b><br/>HODS<br/><b>Order Placer</b>"]
TestType[Test Distribution<br/>By Test Type] --> |4b. Tests A, B, C, etc| GLHS
TestType --> |4b. Tests D, E etc| GLHI
GLHS["<b>Order Filler</b><br/>LIMS Shire"]
GLHS --> |4c. Send Laboratory Report<br/>HL7 v2 ORU_R01| RIE
GLHI["<b>Order Filler</b><br/>LIMS iGene)"]
GLHI --> |4c. Send Laboratory Report<br/>| RIE
end
HODS --> |5. Write Consolidated Report| HODS
HODS --> |"6a. Send Consolidated Laboratory Report<br/>Email"| RIE
RIE --> |6b Send Laboratory Report| TIE
TIE --> |Laboratory Report| EPR
classDef purple fill:#E1D5E7;
class NHSTrust,HODS,GLHS,GLHI,LIMS purple
For information purposes only. This is a more detailed breakdown of the Genomic Tests.
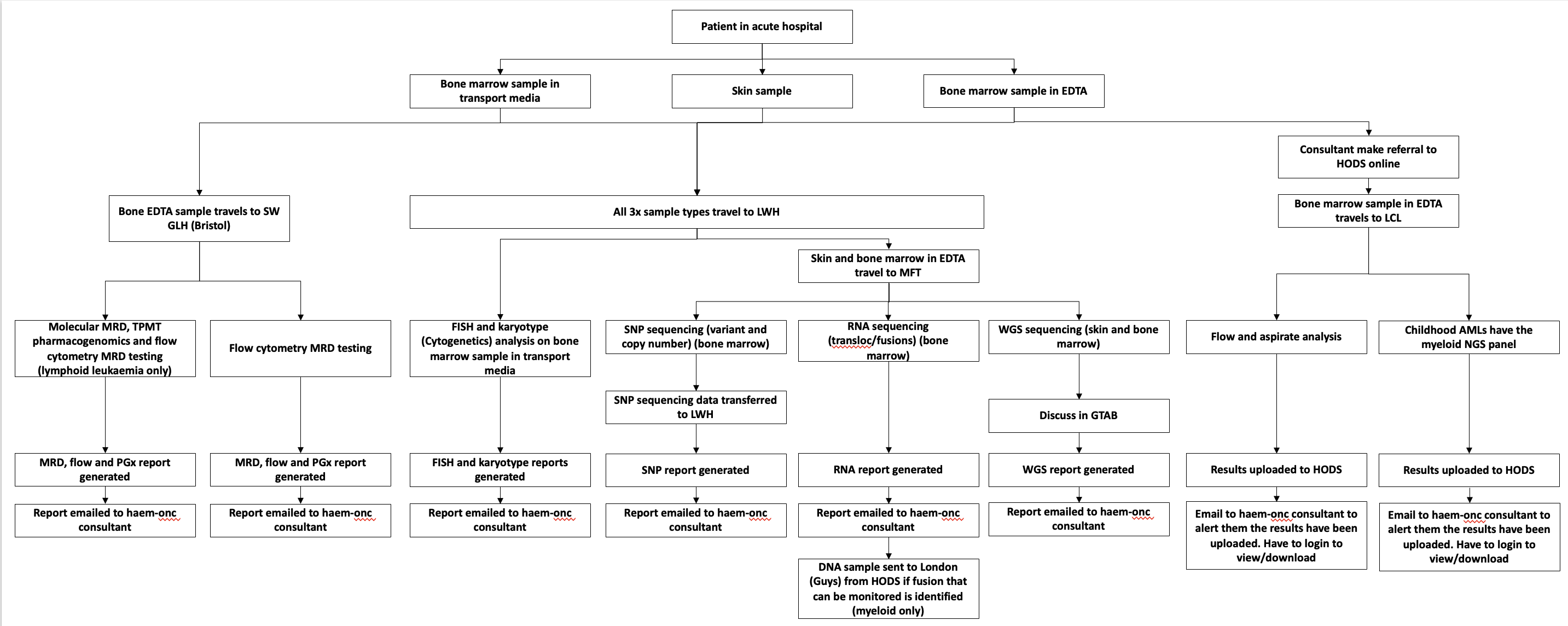
HODS Genomic Tests - Mersey and Cheshire GLH
TBD - Starlims
graph TD;
subgraph NHSTrustA[Cheshire and Mersey NHS Trust]
EPRA[<b>Order Placer</b>] --> |2. Asks For| SpecimenA[Sample Collection]
TIEA["Trust Integration Engine"]
EPRA --> |1a. Laboratory Order| TIEA
end
subgraph NHSTrustB[Greater Manchester NHS Trust]
EPRB[<b>Order Placer</b>] --> |2. Asks For| SpecimenB[Sample Collection]
TIEB[Trust Integration Engine]
EPRB --> |1a. Laboratory Order| TIEB
end
SpecimenA --> |3. Send Specimen| LIMSA
SpecimenB --> |3. Send Specimen| LIMSB
subgraph NWGenomics[NW Genomics]
TIEA --> |1b. Laboratory Order| RIE
TIEB --> |1b. Laboratory Order| RIE
RIE --> |1c. Laboratory Order| LIMSA[<b>Order Filler</b><br/>Liverpool LIMS Starlims]
RIE --> |1c. Laboratory Order| LIMSB[<b>Order Filler</b><br/>Manchester LIMS iGene]
LIMSA <--> |3. Redistribution of Orders and Specimen by test type| LIMSB
LIMSA --> |4a. Laboratory Report| RIE[Regional Integration Engine]
LIMSB --> |4a. Laboratory Report| RIE
end
RIE --> |4b. Laboratory Report| EPRA
RIE --> |4b. Laboratory Report| EPRB
classDef purple fill:#E1D5E7;
class EPRA,EPRB,SpecimenA,SpecimenB,LIMSA,LIMSB purple;
The introduction of the Regional Integration Engine (RIE) creates a single mechanism for sending orders and reports to the appropriate regional laboratories. It abstracts the differing order and reporting formats required by each LIMS, so these variations are hidden from the NHS Trusts.
Orders can be delivered to each LIMS in two ways:
While the Automated Test Order Delivery approach is technically simpler to implement, it may not support effective laboratory order and specimen management. NW Genomics will also receive reflex and subcontracted orders, particularly within Cancer Pathways. Additionally, with the upcoming NHS England Genomic Order Management System (GOMS), reflex and subcontracted orders are expected to increase from other NHS Genomics organisations.
graph TD;
subgraph NHSTrustA[NHS Trust]
EPRA[<b>Order Placer</b>] --> |Asks For| SpecimenA[Sample Collection]
EPRA --> |1a. Laboratory Order| TIE[Trust Integration Engine]
end
SpecimenA --> |2 Send Specimen| LIMSA
SpecimenA --> |2 Send Specimen| LIMSB
SpecimenA --> |2 Send Specimen| LIMSC
SpecimenA --> |2 Send Specimen| LIMSD
TIE --> |1b. Laboratory Order| RIE
subgraph NWGenomics[NW Genomics]
RIE --> |1c. Laboratory Order| TD[Test Distribution<br/>By Test Type]
TD --> |1d. Laboratory Order| LIMSA[<b>Order Filler</b><br/>LIMS iGene]
TD --> |1d. Laboratory Order| LIMSB[<b>Order Filler</b><br/>LIMS Starlims]
TD --> |1d. Laboratory Order| LIMSC[<b>Order Filler</b><br/>LIMS Shire]
TD --> |1d. Laboratory Order| LIMSD[<b>Order Filler</b><br/>LIMS Histotrac]
LIMSA --> |4a. Laboratory Report| RIE[Regional Integration Engine]
LIMSB --> |4a. Laboratory Report| RIE
LIMSC --> |4a. Laboratory Report| RIE
LIMSD --> |4a. Laboratory Report| RIE
end
RIE --> |4b. Laboratory Report| TIE
TIE --> |4c. Laboratory Report| EPRA
classDef purple fill:#E1D5E7;
class EPRA,SpecimenA,LIMSA,LIMSB,LIMSC,LIMSD purple;
See also Inter Laboratory Workflow (ILW)
graph TD;
subgraph NHSTrustA[NHS Trust]
EPRA[<b>Order Placer</b>] --> |Asks For| SpecimenA[Sample Collection]
EPRA --> |1a. Laboratory Order| TIE[Trust Integration Engine]
end
SpecimenA --> |2 Send Specimen| LIMSA
SpecimenA --> |2 Send Specimen| LIMSB
SpecimenA --> |2 Send Specimen| LIMSC
SpecimenA --> |2 Send Specimen| LIMSD
TIE --> |1b. Laboratory Order<br/>LAB-1| RIE
subgraph NWGenomics[NW Genomics]
RIE --> |1c. Laboratory Order<br/>LAB-1| LIMSA[<b>Order Filler</b><br/>LIMS iGene]
LIMSA --> |1d. Subcontracted Laboratory Order<br/>LAB-35| LIMSB[<b>Order Filler</b><br/>LIMS Starlims]
LIMSA --> |1d. Subcontracted Laboratory Order<br/>LAB-35| LIMSC[<b>Order Filler</b><br/>LIMS Shire]
LIMSA --> |1d. Subcontracted Laboratory Order<br/>LAB-35| LIMSD[<b>Order Filler</b><br/>LIMS Histotrac]
LIMSA --> |4a. Laboratory Report| RIE[Regional Integration Engine]
LIMSB --> |4a. Laboratory Report| RIE
LIMSC --> |4a. Laboratory Report| RIE
LIMSD --> |4a. Laboratory Report| RIE
end
RIE --> |4b. Laboratory Report| TIE
TIE --> |4c. Laboratory Report| EPRA
classDef purple fill:#E1D5E7;
class EPRA,SpecimenA,LIMSA,LIMSB,LIMSC,LIMSD purple;
graph TD;
subgraph NHSTrust[NHS Trust]
Practitioner[fas:fa-user-md Practitioner] --> |1. Selects Order Form| FormManager
FormManager --> OrderEntry
Practitioner --> |3. Completes| OrderEntry[Order Form]
EPR[<b>Order Placer</b><br/>fas:fa-database Electronic Patient Record] --> |2. Pre Populates with existing data| OrderEntry
OrderEntry --> |4. Submits Order| EPR
Practitioner --> |6. Asks for|Sample[Sample Collection]
end
EPR --> |5. Sends Laboratory Order<br/>LAB-1 HL7 FHIR Message O21| DiagnosticTesting[<b>Order Filler</b><br/>fas:fa-stethoscope Diagnostic Testing]
Sample --> DiagnosticTesting
For more details see:
graph TD;
Sample[Sample Collection] --> EXT
Order --> EXT
subgraph OrderFiller[<b>Order Filler</b> North West Genomics]
EXT[DNA Extraction] --> SEQ[DNA Sequencing]
SEQ --> AN[Mapping & Analysis]
AN --> INT[Interpretation]
end
INT --> |Send Laboratory Report<br/>LAB-3 HL7 v2 ORU_R01| Practitioner[<b>Order Placer</b><br/>EPR]
For more details see:
For illustration purposes only, see Inter Laboratory Workflow
For illustration purposes only, see Specimen Event Tracking