Genomics Reporting Implementation Guide, published by HL7 International / Clinical Genomics. This guide is not an authorized publication; it is the continuous build for version 4.0.0-ballot built by the FHIR (HL7® FHIR® Standard) CI Build. This version is based on the current content of https://github.com/HL7/genomics-reporting/ and changes regularly. See the Directory of published versions
| Official URL: http://hl7.org/fhir/uv/genomics-reporting/OperationDefinition/find-subject-molecular-consequences | Version: 4.0.0-ballot | |||
| Standards status: Trial-use | Maturity Level: 2 | Computable Name: FindSubjectMolecularConsequences | ||
Retrieves molecular consequences of a DNA variant.
Retrieves molecular consequences of a DNA variant.
Generally, the operation will be called with 'subject' and 'ranges' parameters, which will return molecular consequences for patient variants in supplied ranges. Another common pattern is to call the operation with 'subject', 'ranges', and 'featureConsequences' parameters, which will limit the return to only those molecular consequences having supplied features. For other combinations of input parameters, the operation returns molecular consequences satisfying all parameters.
Where no variants, ranges, or featureConsequences are provided, the operation returns no consequences.
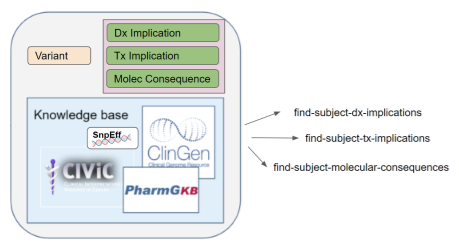
As shown in the following picture, this operation can return previously instantiated implications and/or dynamically computed implications. Specific implementations can indicate their capabilities using a FHIR Capability Statement. Rules around the retention of dynamically computed implications are outside the scope of this operation, but a server could potentially instantiate those results based on the Therapeutic Implication, Diagnostic Implication, or Molecular Consequence FHIR profiles.
| Use | Name | Scope | Cardinality | Type | Binding | Documentation |
| IN | subject | 1..1 | string (reference) | The subject of interest. | ||
| IN | variants | 0..* | string (string) | List of variants from which consequences are derived. Must be in HGVS or SPDI format. | ||
| IN | ranges | 0..* | string (special) | List of regions to be searched for variants. Must be in zero-based RefSeq:Integer-range format (e.g. 'NC_000007.14:55174721-55174820'). | ||
| IN | featureConsequences | 0..* | string (token) | List of consequences sought. Must be in token or codesystem|code format. (These will generally be coded with Sequence Ontology codes under SO:0001537) | ||
| IN | testIdentifiers | 0..* | string (token) | Supply a list of test identifiers. Only results originating from one of these tests will be returned. | ||
| IN | testDateRange | 0..1 | Period | Supply a date range. Only results generated during this range will be returned. | ||
| IN | specimenIdentifiers | 0..* | string (token) | Supply a list of specimen identifiers. Only results derived from one of these specimens will be returned. | ||
| IN | genomicSourceClass | 0..1 | string (token) | Enables an App to limit results to those that are 'germline' or 'somatic'. Default is to include variants irrespective of genomic source class. | ||
| OUT | consequence | 0..* | Observation | Consequences must conform to [Profile: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/molecular-consequence]. The target(s) of the consequence's derivedFrom shall also be included in the response. | ||
| OUT | variant | 0..* | Observation | Variants must conform to [Profile: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/variant]. |
Valid response codes are shown in the following table. Additional response codes (e.g. 5xx server error) may also be encountered.
| Response Code | Description |
|---|---|
| 200 | Successfully executed request |
| 400 | ERROR: Invalid query parameters |
| 404 | ERROR: Patient not found |
| 422 | ERROR: Failed LiftOver |
A patient with lung cancer has no known druggable targets. Their clinician wants to see if any variants in ALK (NC_000002.12:29190991-29921589), BRAF (NC_000007.14:140713327-140924929), or EGFR (NC_000007.14:55019016-55211628) are predicted to be consequential.
$find-subject-molecular-consequences?subject=HG00403&ranges=NC_000002.12:29190991-29921589,NC_000007.14:140713327-140924929,NC_000007.14:55019016-55211628
The clinician wants to further constrain the query in the prior example to only return variants predicted to affect splice donor sites
$find-subject-molecular-consequences?subject=HG00403&ranges=NC_000002.12:29190991-29921589,NC_000007.14:140713327-140924929,NC_000007.14:55019016-55211628&featureConsequences=http://www.sequenceontology.org|SO:0001575
Patient HG00403 is detected as having a variant (NC_000001.10:86852620:A:G) in the ODF2L gene, and their clinician wants to further characterize the variant by examining its predicted molecular consequences. (Results indicate that the variant affects a splice donor site and can lead to gene loss of function).