MedMorph Research Data Exchange Content IG, published by HL7 International - Public Health Work Group. This is not an authorized publication; it is the continuous build for version 0.1.0). This version is based on the current content of https://github.com/HL7/fhir-medmorph-research-dex-ig/ and changes regularly. See the Directory of published versions
The section identifies the business needs and specific user stories outlining the research data exchange needs.
The purpose of the Making Electronic Data More Available for Research and Public Health (MedMorph) Research Data Exchange Implementation Guide (IG) is to streamline and expedite the process of onboarding research data partners and contributing data to research networks. The current processes used to extract, transform, and load (ETL) the data and then contribute the data involve many non-standardized mechanisms (e.g., Secure File Transfer Protocol (SFTP), Excel Files, stored procedures), different structures (e.g., formats), and different semantics. As a result of these non-standardized processes, the length of time to onboard a data partner varies from weeks to months. This research data exchange use case along with leveraging the MedMorph Reference Architecture (RA) IG with other existing Health Level 7 (HL7®) Fast Healthcare Interoperability Resources (FHIR®) IGs will help reduce the length of time it takes to onboard new data exchange partners.
The goals of the Research Data Exchange use case include:
In Scope
Out of Scope
As a research network administrator, there is a need to onboard research data partners to join the research network and contribute data that can be used for research. The data partners extract, transform, and load (ETL) the data from data sources such as Electronic Health Records (EHRs), Health Information Exchanges (HIEs), or other data repositories (e.g., clinical data warehouses). These processes (e.g., ETL) when standardized will expedite onboarding processes and deliver better quality data at a lower cost.
Once a data partner joins a research network and contributes data, a researcher should be able to perform queries to filter specific data sets and analyze the data. Currently researchers can query data marts on a limited basis because of the variations in the data models (e.g., National Patient-Centered Clinical Research Network (PCORnet) Clinical Data Management (CDM), Informatics for Integrating Biology & the Bedside (i2b2), Observational Medical Outcomes Partnership (OMOP), FHIR Resources) and the technologies (e.g., Microsoft Structured Query Language (SQL), Postgres SQL, Statistical Analysis System (SAS)) used to host the data mart. Standardizing these data access mechanisms will help overcome the hurdles faced by researchers in accessing data from data partners and EHRs. NOTE: User Story #2 is out of scope for this version of the IG, it is being included only as a potential use case that could be expanded on in the future.
The focus of the MedMorph Research Data Exchange IG is User Story #1. This is based on extensive discussions with the PCORnet Front Door team, the MedMorph Reference Architecture (RA) WG, and the CDC MedMorph team. The reasons for choosing User Story #1 is as follows:
The current state is as shown in the Figure below:
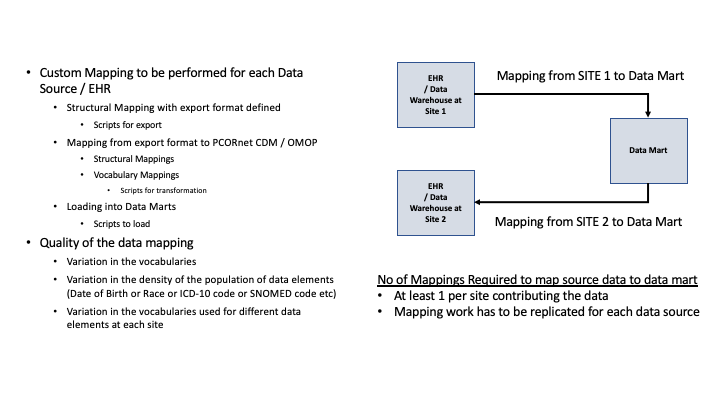
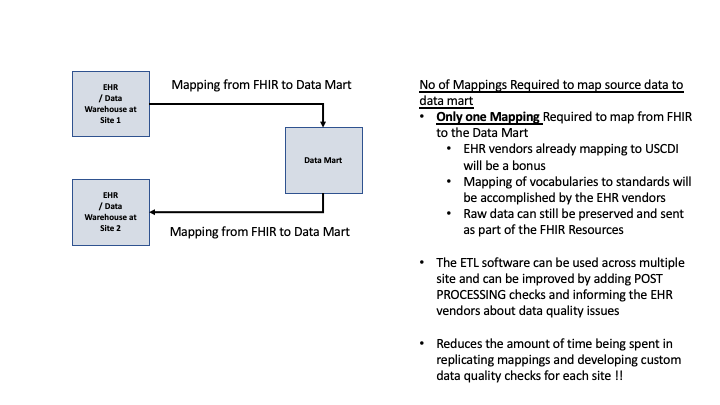
The usage of FHIR during the onboarding process and expected efficiencies are documented below in Figure 2.2.
The following actors from the MedMorph RA IG are used by the Research Data Exchange use case:
Figure 2.3 below shows the research abstract model to onboard a data partner who can contribute data from their data source (e.g., and EHR system) to populate a data mart.
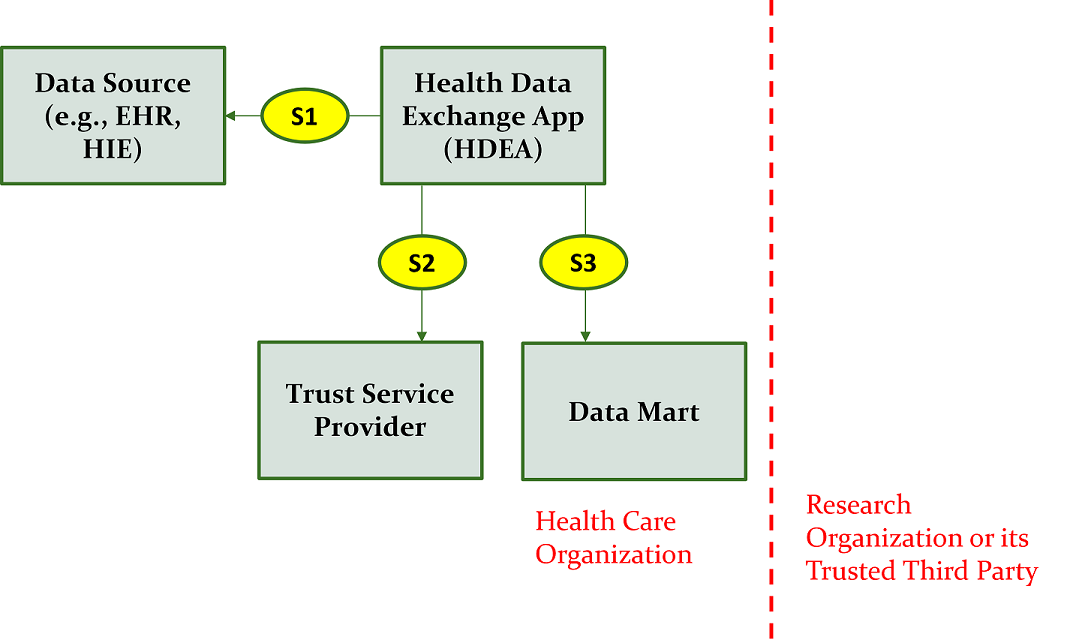
The description of the interactions as illustrated in Figure 2.3 include:
As an alternative to Figure 2.3, the Data Mart and the HDEA could reside outside the health care organization as shown in Figure 2.4 below.
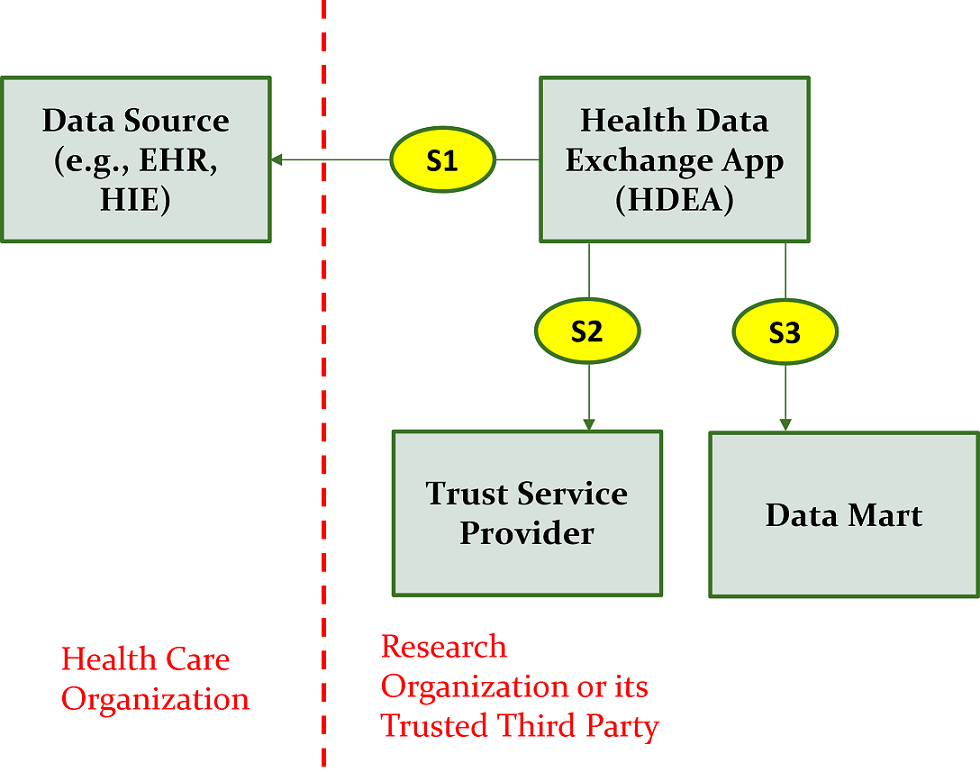
The description of the interactions as illustrated in Figure 2.4 include: