MedMorph Research Data Exchange Content IG, published by HL7 International - Public Health Work Group. This is not an authorized publication; it is the continuous build for version 0.1.0). This version is based on the current content of https://github.com/HL7/fhir-medmorph-research-dex-ig/ and changes regularly. See the Directory of published versions
This section defines the specific requirements for systems wishing to conform to actors specified in this Making Electronic Data More Available for Research and Public Health (MedMorph) Research Data Exchange Implementation Guide (IG). The specification focuses on using the Health Data Exchange App (HDEA), MedMorph’s backend services app, to perform the extract, transform, and load (ETL) process used for data partner on boarding within research networks.
Before reading this formal specification, implementers should first be familiar with the Use Cases page which provides the business context and general process flow.
This IG uses specific terminology to flag statements that have relevance for the evaluation of conformance with the guide:
SHALL indicates requirements that must be met to be conformant with the specification.
SHOULD indicates behaviors that are strongly recommended (and which may result in interoperability issues or sub-optimal behavior if not adhered to), but which do not, for this version of the specification, affect the determination of specification conformance.
MAY describes optional behaviors that are free to consider but where there is no recommendation for or against adoption.
Actors and Systems asserting conformance to this IG must implement the requirements outlined in the corresponding capability statements. The following definition of Must Support is to be used in the implementation of the requirements.
This specification makes significant use of FHIR profiles, search parameter definitions, and terminology artifacts to describe the content to be shared as part of MedMorph Research Data Exchange IG workflows. The IG is based on FHIR R4 and profiles are listed for each interaction.
The full set of profiles defined in this IG can be found by following the links on the FHIR Artifacts page.
This IG leverages the MedMorph RA IG published by HL7 Public Health Work Group (WG) as the reference architecture for automation and implementing the research data exchange use case.
This IG leverages the CDMH IG profiles to export and map data that can be transformed to National Patient-Centered Clinical Research Network (PCORnet) Common Data Model (CDM), Informatics for Integrating Biology and the Bedside (i2b2), Observational Medical Outcomes Partnership (OMOP), and Sentinel data models. The CDMH profiles provide a complete set of mapping from FHIR profiles to PCORnet CDM which will be leveraged by the implementers of this IG to transform the FHIR resources to the appropriate CDM hosted by the Data Mart.
This IG leverages the US Core set of profiles defined by HL7 for sharing non-veterinary EMR individual health data in the U.S. Where US Core profiles exist, this IG either leverages them directly or uses them as a base for any additional constraints needed to support the research use cases. If no constraints are needed, this IG does not define any profiles.
This IG leverages the BulkData Access IG published by the HL7 FHIR Infrastructure WG for:
* Performing a Bulk Export of data (e.g., Group Export Operation) from a Data Source that can be used to populate the Data Mart CDM.
* SMART on FHIR Backend Services for Authorization
This section describes the requirements of the Research Knowledge Artifact to implement the outlined use cases. In order to define the Knowledge Artifact, the following workflow is used.
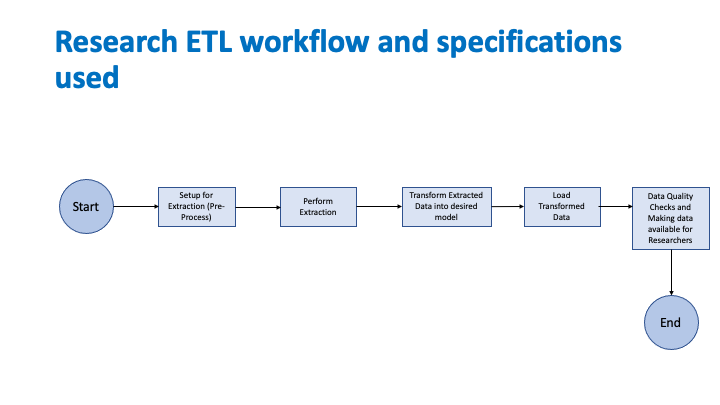
The Knowledge Artifact should have appropriate actions for data extraction from a Data Source, data transformation to the appropriate CDM, and data load into the Data Mart CDM. However, the MedMorph RA IG currently defines actions only to extract the data from the Data Source but does not provide generic actions for transforming and loading the data into the data mart. The Knowledge Artifact for the Research Data Exchange will contain the actions to extract the data but not the actions for transformation and loading the data mart.
This section outlines how the SMART on FHIR Backend Services Authorization from the Bulk Data Access IG will be used by the MedMorph Research Data Exchange IG.
The system actors namely the Data Source and the HDEA are required to use the SMART on FHIR Backend Services Authorization mechanisms as outlined below for the following interactions
System actors acting as servers (e.g., electronic health record (EHR)) SHALL advertise conformance to SMART Backend Services by hosting a Well-Known Uniform Resource Identifiers (URIs) as defined in the Bulk Data Access IG Authorization Section specification.
System actors acting as servers SHALL include token_endpoint, scopes_supported, token_endpoint_auth_methods_supported and token_endpoint_auth_signing_alg_values_supported as defined in the Bulk Data Access IG Authorization Section specification.
When System actors act as clients (e.g., HDEA), they SHALL share their JSON Web Key Set (JWKS) with the server System actors (e.g., Data Source) using Uniform Resource Locators (URLs) as defined in the Bulk Data Access IG Authorization Section specification.
System actors acting as clients SHALL obtain the access token as defined in the Bulk Data Access IG Authorization Section specification.
For the research data exchange use cases, Data Sources SHALL support the system/*.read scopes.
The healthcare organization’s existing processes along with the Data Source’s authorization server SHALL verify consent and other policy requirements before allowing the HDEA to access the data to be extracted for research purposes. The processes to be verified by the health care organizations include any consent required from patients, Institutional Review Board (IRB) approvals and any other agreements required based on the health care organization policies.
The Data Source MAY support the creation of Group resource to identify the patients who have consented to be participating in a research program.
The Data Source SHALL associate Groups with specific clients who can query the Group for data.
The Data Source MAY allow clients to retrieve Group resource by name. When multiple Group resources meet the criteria, the Data Source MAY return each of the Group resources that match the name, unless the Data Source can narrow down the Group resource to only one.
The Data Source MAY allow clients to retrieve Group resource by identifier. When multiple Group resources meet the criteria, the Data Source MAY return each of the Group resources that match the name, unless the Data Source can narrow down the Group resource to only one.
The Data Source and the HDEA SHOULD pre-coordinate the group names and/or identifiers that can be used for retrieval of data.
The Data Source SHALL support the Bulk Data Access Group export operation to export the data required for the data mart population.
The Data Source SHALL support the Group export operation parameters for all Group export operations.
The Data Source SHALL support the US Core Profiles as outlined in the Data Source Capability Statement for the HDEA to access patient data using the Group export operation.
The Data Source SHOULD support the CDMH IG Profiles for the HDEA to access patient data using the Group export operation for populating multiple common data models.
The HDEA SHALL allow the healthcare organization to activate/deactivate a specific Knowledge Artifact. Activation indicates applying the Knowledge Artifact and deactivation indicates not applying the Knowledge Artifact for events occurring within the healthcare organization.
The HDEA SHALL implement FHIRPath expression processing to process the MedMorph Research Data Exchange Knowledge Artifact actions.
The HDEA SHALL schedule jobs to perform data extraction from Data Sources at scheduled time intervals per the Knowledge Artifact.
The HDEA acting as a client SHALL use the US Core Server APIs as outlined in the Data Source Capability Statement to access patient data from the Data Source.
The HDEA acting as a client SHOULD use the CDMH IG Profiles to extract data required to populate the common data models.
ndjson to appropriate data models and load the data models following the CDMH IG Mappings from FHIR to CDMs.