minimal Common Oncology Data Elements (mCODE) Implementation Guide, published by HL7 International / Clinical Interoperability Council. This guide is not an authorized publication; it is the continuous build for version 4.0.0-ballot built by the FHIR (HL7® FHIR® Standard) CI Build. This version is based on the current content of https://github.com/HL7/fhir-mCODE-ig/ and changes regularly. See the Directory of published versions
| Official URL: http://hl7.org/fhir/us/mcode/StructureDefinition/mcode-genomic-variant | Version: 4.0.0-ballot | |||
| Active as of 2024-04-03 | Maturity Level: 1 | Computable Name: GenomicVariant | ||
| Other Identifiers: OID:2.16.840.1.113883.4.642.40.15.42.19 | ||||
Details about a set of changes in the tested sample compared to a reference sequence. The term variant can be used to describe an alteration that may be benign, pathogenic, or of unknown significance. The term variant is increasingly being used in place of the term mutation. Variants can be computed relative to reference sequence assembly from which it was identified.
GenomicVariant SHOULD be sent as part of a GenomicsReport and MAY be sent as a standalone observation.
Observation resources associated with an in-scope patient with Observation.code LOINC 69548-6 SHALL conform to this profile. Beyond this requirement, a producer of resources SHOULD ensure that any resource instance associated with an in-scope patient that would reasonably be expected to conform to this profile SHOULD be published in this form.
Usage:
Changes since version true:
Description of Profiles, Differentials, Snapshots and how the different presentations work.
This structure is derived from Variant
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | 0..* | Variant | Variant | |
  | S | 1..1 | code | registered | preliminary | final | amended + |
  | S | 1..* | CodeableConcept | Classification of type of observation |
  | 1..1 | CodeableConcept | Classification of type of observation Required Pattern: At least the following | |
   | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
    | 1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |
    | 1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |
   | 1..1 | Coding | Code defined by a terminology system | |
  | S | 1..1 | CodeableConcept | 69548-6 |
  | S | 0..1 | Reference(Cancer Patient Profile) | Who and/or what the observation is about |
  | S | 0..1 | dateTime, Period, Timing, instant | Clinically relevant time/time-period for observation Slice: Unordered, Open by type:$this |
   | S | 0..1 | dateTime | Clinically relevant time/time-period for observation |
  | S | 0..1 | CodeableConcept | Actual result |
  | S | 0..1 | CodeableConcept | Why the result is missing |
  | S | 0..1 | CodeableConcept | Sequencing | SNP array | PCR | Computational analysis | ... |
  | S | 0..1 | Reference(Human Specimen Profile) | Specimen used for this observation |
  | 0..* | BackboneElement | Not used in this profile | |
  | 0..* | Reference(Observation | QuestionnaireResponse | MolecularSequence) | Not used in this profile | |
  | S | 0..* | BackboneElement | Component results |
  | S | 0..* | BackboneElement | Gene Studied |
   | S | 1..1 | CodeableConcept | 48018-6 |
   | S | 1..1 | CodeableConcept | The HGNC gene symbol is to be used as display text and the HGNC gene ID used as the code. If no HGNC code issued for this gene yet, NCBI gene IDs SHALL be used. |
  | S | 0..* | BackboneElement | Cytogenetic (Chromosome) Location |
   | S | 1..1 | CodeableConcept | 48001-2 |
   | S | 1..1 | CodeableConcept | Example: 1q21.1 |
  | S | 0..1 | BackboneElement | Genomic (gDNA) Change - gHGVS |
   | S | 1..1 | CodeableConcept | 81290-9 |
   | S | 1..1 | CodeableConcept | A valid HGVS-formatted 'g.' string, e.g. NC_000016.9:g.2124200_2138612dup |
  | S | 0..1 | BackboneElement | Cytogenomic Nomenclature (ISCN) |
   | S | 1..1 | CodeableConcept | 81291-7 |
   | S | 1..1 | CodeableConcept | Actual component result |
  | S | 0..1 | BackboneElement | Coding DNA Change Type |
   | S | 1..1 | CodeableConcept | 48019-4 |
   | S | 1..1 | CodeableConcept | deletion | insertion | delins | SNV | copy_number_gain | copy_number_loss | ... (many) |
  | S | 0..1 | BackboneElement | Genomic Source Class |
   | S | 1..1 | CodeableConcept | 48002-0 |
   | S | 1..1 | CodeableConcept | Germline | Somatic | Fetal | Likely germline | Likely somatic | Likely fetal | Unknown genomic origin | De novo |
  | S | 0..1 | BackboneElement | Sample Allelic Frequency |
   | S | 1..1 | CodeableConcept | 81258-6 |
   | S | 0..1 | Quantity | Relative frequency in the sample |
  | S | 0..1 | BackboneElement | Allelic State |
   | S | 1..1 | CodeableConcept | 53034-5 |
   | S | 1..1 | CodeableConcept | Heteroplasmic | Homoplasmic | Homozygous | Heterozygous | Hemizygous |
  | S | 0..* | BackboneElement | Variation Code |
   | S | 1..1 | CodeableConcept | 81252-9 |
   | S | 1..1 | CodeableConcept | ClinVar ID or similar |
  | S | 0..1 | BackboneElement | Protein (Amino Acid) Change - pHGVS |
   | S | 1..1 | CodeableConcept | 48005-3 |
   | S | 1..1 | CodeableConcept | A valid HGVS-formatted 'p.' string, e.g. NP_000050.2:p.(Asn1836Lys) |
  | S | 0..1 | BackboneElement | Amino Acid Change Type |
   | S | 1..1 | CodeableConcept | 48006-1 |
   | S | 1..1 | CodeableConcept | Wild type | Deletion | Duplication | Frameshift | Initiating Methionine | Insertion | Insertion and Deletion | Missense | Nonsense | Silent |
  | S | 0..1 | BackboneElement | Molecular Consequence |
   | S | 1..1 | CodeableConcept | molecular-consequence |
   | S | 1..1 | CodeableConcept | stop_lost | stop_gained | inframe_insertion | frameshift_variant | ... (many) |
  | S | 0..1 | BackboneElement | Genomic Structural Variant Copy Number |
   | S | 1..1 | CodeableConcept | 82155-3 |
   | S | 0..1 | Quantity | Actual component result |
 Documentation for this format Documentation for this format | ||||
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | C | 0..* | Variant | Variant obs-6: dataAbsentReason SHALL only be present if Observation.value[x] is not present obs-7: If Observation.code is the same as an Observation.component.code then the value element associated with the code SHALL NOT be present |
  | ?!Σ | 0..1 | uri | A set of rules under which this content was created |
  | 0..1 | CodeableConcept | Secondary findings are genetic test results that provide information about variants in a gene unrelated to the primary purpose for the testing, most often discovered when [Whole Exome Sequencing (WES)](https://en.wikipedia.org/wiki/Exome_sequencing) or [Whole Genome Sequencing (WGS)](https://en.wikipedia.org/wiki/Whole_genome_sequencing) is performed. This extension should be used to denote when a genetic finding is being shared as a secondary finding, and ideally refer to a corresponding guideline or policy statement.
For more detail, please see:
https://ghr.nlm.nih.gov/primer/testing/secondaryfindings URL: http://hl7.org/fhir/StructureDefinition/observation-secondaryFinding Binding: GeneticObservationSecondaryFindings (extensible): Codes to denote a guideline or policy statement when a genetic test result is being shared as a secondary finding. | |
  | 0..1 | Reference(BodyStructure) | Target anatomic location or structure URL: http://hl7.org/fhir/StructureDefinition/bodySite | |
  | ?! | 0..* | Extension | Extensions that cannot be ignored |
  | ?!SΣ | 1..1 | code | registered | preliminary | final | amended + Binding: ObservationStatus (required): Codes providing the status of an observation. |
  | S | 1..* | CodeableConcept | Classification of type of observation Slice: Unordered, Open by value:coding Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. |
   | 1..1 | CodeableConcept | Classification of type of observation Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. Required Pattern: At least the following | |
    | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |
    | Σ | 1..1 | Coding | Code defined by a terminology system Required Pattern: At least the following |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |
  | SΣ | 1..1 | CodeableConcept | 69548-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
   | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
    | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
    | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 69548-6 | |
  | SΣ | 0..1 | Reference(Cancer Patient Profile) | Who and/or what the observation is about |
  | SΣ | 0..1 | Clinically relevant time/time-period for observation Slice: Unordered, Open by type:$this | |
   | dateTime | |||
   | Period | |||
   | Timing | |||
   | instant | |||
   | SΣ | 0..1 | dateTime | Clinically relevant time/time-period for observation |
  | SΣC | 0..1 | CodeableConcept | Actual result Slice: Unordered, Closed by type:$this |
  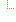 | ΣC | 0..1 | CodeableConcept | Indeterminate | No call | Present | Absent. Binding: LOINC Answer List LL1971-2 (required) |
  | SC | 0..1 | CodeableConcept | Why the result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
  | S | 0..1 | CodeableConcept | Sequencing | SNP array | PCR | Computational analysis | ... Binding: LOINC Answer List LL4048-6 (extensible) |
  | S | 0..1 | Reference(Human Specimen Profile) | Specimen used for this observation |
  | C | 0..* | BackboneElement | Not used in this profile obs-3: Must have at least a low or a high or text |
   | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
  | Σ | 0..* | Reference(Observation | QuestionnaireResponse | MolecularSequence) | Not used in this profile |
  | SΣ | 0..* | BackboneElement | Component results Slice: Unordered, Open by pattern:code |
   | Content/Rules for all slices | |||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
   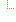 | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. |
   | Σ | 0..1 | BackboneElement | Clinical Conclusion |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | conclusion-string Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: conclusion-string | |
   | SΣ | 0..* | BackboneElement | Gene Studied |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 48018-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48018-6 | |
    | SΣ | 1..1 | CodeableConcept | The HGNC gene symbol is to be used as display text and the HGNC gene ID used as the code. If no HGNC code issued for this gene yet, NCBI gene IDs SHALL be used. Binding: HUGO Gene Nomenclature Committee Gene Names (HGNC) (extensible) |
   | SΣ | 0..* | BackboneElement | Cytogenetic (Chromosome) Location |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 48001-2 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48001-2 | |
    | SΣ | 1..1 | CodeableConcept | Example: 1q21.1 |
   | Σ | 0..* | BackboneElement | Human Reference Sequence Assembly |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 62374-4 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 62374-4 | |
    | Σ | 1..1 | CodeableConcept | GRCh37 | GRCh38 | ... Binding: LOINC Answer List LL1040-6 (extensible) |
   | Σ | 0..1 | BackboneElement | Coding (cDNA) Change - cHGVS |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 48004-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48004-6 | |
    | Σ | 1..1 | CodeableConcept | A valid HGVS-formatted 'c.' string, e.g. NM_005228.5:c.2369C>T. Binding: Human Genome Variation Society (HGVS) Nomenclature (required) |
   | SΣ | 0..1 | BackboneElement | Genomic (gDNA) Change - gHGVS |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 81290-9 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81290-9 | |
    | SΣ | 1..1 | CodeableConcept | A valid HGVS-formatted 'g.' string, e.g. NC_000016.9:g.2124200_2138612dup Binding: Human Genome Variation Society (HGVS) Nomenclature (required) |
   | SΣ | 0..1 | BackboneElement | Cytogenomic Nomenclature (ISCN) |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 81291-7 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81291-7 | |
    | SΣ | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined |
   | Σ | 0..1 | BackboneElement | Genomic Reference Sequence |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 48013-7 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48013-7 | |
    | Σ | 1..1 | CodeableConcept | Versioned genomic reference sequence identifier Binding: (unbound) (example): Multiple bindings acceptable (NCBI or LRG) |
   | Σ | 0..1 | BackboneElement | Reference Transcript |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 51958-7 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 51958-7 | |
    | Σ | 1..1 | CodeableConcept | Versioned transcript reference sequence identifier Binding: (unbound) (example): Multiple bindings acceptable (NCBI or LRG) |
   | Σ | 0..1 | BackboneElement | Exact Start-End |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 81254-5 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81254-5 | |
   | Σ | 0..1 | BackboneElement | Inner Start-End |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 81302-2 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81302-2 | |
   | Σ | 0..1 | BackboneElement | Outer Start-End |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 81301-4 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81301-4 | |
   | Σ | 0..1 | BackboneElement | Coordinate System |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 92822-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 92822-6 | |
    | Σ | 1..1 | CodeableConcept | 0-based interval counting | 0-based character counting | 1-based character counting Binding: LOINC Answer List LL5323-2 (extensible) |
   | Σ | 0..1 | BackboneElement | Genomic Ref Allele |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 69547-8 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 69547-8 | |
   | Σ | 0..1 | BackboneElement | Genomic Alt Allele |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 69551-0 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 69551-0 | |
   | SΣ | 0..1 | BackboneElement | Coding DNA Change Type |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 48019-4 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48019-4 | |
    | SΣ | 1..1 | CodeableConcept | deletion | insertion | delins | SNV | copy_number_gain | copy_number_loss | ... (many) Binding: DNA Change Type (extensible): Concepts in sequence ontology under SO:0002072 |
   | SΣ | 0..1 | BackboneElement | Genomic Source Class |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 48002-0 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48002-0 | |
    | SΣ | 1..1 | CodeableConcept | Germline | Somatic | Fetal | Likely germline | Likely somatic | Likely fetal | Unknown genomic origin | De novo Binding: LOINC Answer List LL378-1 (extensible) |
   | SΣ | 0..1 | BackboneElement | Sample Allelic Frequency |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 81258-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81258-6 | |
    | SΣ | 0..1 | Quantity | Relative frequency in the sample |
     | ?!Σ | 0..1 | code | < | <= | >= | > - how to understand the value Binding: QuantityComparator (required): How the Quantity should be understood and represented. |
   | Σ | 0..1 | BackboneElement | Allelic Read Depth |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 82121-5 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 82121-5 | |
   | SΣ | 0..1 | BackboneElement | Allelic State |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 53034-5 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 53034-5 | |
    | SΣ | 1..1 | CodeableConcept | Heteroplasmic | Homoplasmic | Homozygous | Heterozygous | Hemizygous Binding: LOINC Answer List LL381-5 (extensible) |
   | Σ | 0..1 | BackboneElement | Variant Inheritance |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | variant-inheritance Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: variant-inheritance | |
    | Σ | 1..1 | CodeableConcept | Maternal | Paternal | Unknown Binding: Variant Inheritances (extensible) |
   | SΣ | 0..* | BackboneElement | Variation Code |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 81252-9 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81252-9 | |
    | SΣ | 1..1 | CodeableConcept | ClinVar ID or similar Binding: (unbound) (example): Multiple bindings acceptable |
   | Σ | 0..* | BackboneElement | Chromosome Identifier |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 48000-4 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48000-4 | |
    | Σ | 1..1 | CodeableConcept | Chromosome 1 | Chromosome 2 | ... | Chromosome 22 | Chromosome X | Chromosome Y Binding: LOINC Answer List LL2938-0 (required) |
   | SΣ | 0..1 | BackboneElement | Protein (Amino Acid) Change - pHGVS |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 48005-3 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48005-3 | |
    | SΣ | 1..1 | CodeableConcept | A valid HGVS-formatted 'p.' string, e.g. NP_000050.2:p.(Asn1836Lys) Binding: Human Genome Variation Society (HGVS) Nomenclature (required) |
   | SΣ | 0..1 | BackboneElement | Amino Acid Change Type |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 48006-1 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48006-1 | |
    | SΣ | 1..1 | CodeableConcept | Wild type | Deletion | Duplication | Frameshift | Initiating Methionine | Insertion | Insertion and Deletion | Missense | Nonsense | Silent Binding: LOINC Answer List LL380-7 (extensible) |
   | SΣ | 0..1 | BackboneElement | Molecular Consequence |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | molecular-consequence Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: molecular-consequence | |
    | SΣ | 1..1 | CodeableConcept | stop_lost | stop_gained | inframe_insertion | frameshift_variant | ... (many) Binding: Molecular Consequence (extensible): Concepts in sequence ontology under SO:0001537. |
   | SΣ | 0..1 | BackboneElement | Genomic Structural Variant Copy Number |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 82155-3 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 82155-3 | |
    | SΣC | 0..1 | Quantity | Actual component result cnt-3: There SHALL be a code with a value of '1' if there is a value. If system is present, it SHALL be UCUM. If present, the value SHALL be a whole number. |
   | Σ | 0..1 | BackboneElement | Variant Confidence Status |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | variant-confidence-status Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: variant-confidence-status | |
    | Σ | 1..1 | CodeableConcept | High | Intermediate | Low Binding: Variant Confidence Status (required) |
 Documentation for this format Documentation for this format | ||||
| Path | Conformance | ValueSet / Code | URI |
| Observation.status | required | ObservationStatushttp://hl7.org/fhir/ValueSet/observation-status|4.0.1from the FHIR Standard | |
| Observation.category | preferred | ObservationCategoryCodeshttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | |
| Observation.category:labCategory | preferred | Pattern: laboratoryhttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | |
| Observation.code | example | Pattern: LOINC code 69548-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.value[x]:valueCodeableConcept | required | LOINC LL1971-2http://loinc.org/vs/LL1971-2 | |
| Observation.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | |
| Observation.method | extensible | LOINC LL4048-6http://loinc.org/vs/LL4048-6 | |
| Observation.component.code | example | LOINCCodes (a valid code from LOINC)http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:conclusion-string.code | example | Pattern: conclusion-stringhttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:gene-studied.code | example | Pattern: LOINC code 48018-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:gene-studied.value[x] | extensible | HGNCVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/hgnc-vs | |
| Observation.component:cytogenetic-location.code | example | Pattern: LOINC code 48001-2http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:reference-sequence-assembly.code | example | Pattern: LOINC code 62374-4http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:reference-sequence-assembly.value[x] | extensible | LOINC LL1040-6http://loinc.org/vs/LL1040-6 | |
| Observation.component:coding-hgvs.code | example | Pattern: LOINC code 48004-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:coding-hgvs.value[x] | required | HGVSVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/hgvs-vs | |
| Observation.component:genomic-hgvs.code | example | Pattern: LOINC code 81290-9http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:genomic-hgvs.value[x] | required | HGVSVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/hgvs-vs | |
| Observation.component:cytogenomic-nomenclature.code | example | Pattern: LOINC code 81291-7http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:cytogenomic-nomenclature.value[x] | example | ||
| Observation.component:genomic-ref-seq.code | example | Pattern: LOINC code 48013-7http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:genomic-ref-seq.value[x] | example | ||
| Observation.component:transcript-ref-seq.code | example | Pattern: LOINC code 51958-7http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:transcript-ref-seq.value[x] | example | ||
| Observation.component:exact-start-end.code | example | Pattern: LOINC code 81254-5http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:inner-start-end.code | example | Pattern: LOINC code 81302-2http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:outer-start-end.code | example | Pattern: LOINC code 81301-4http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:coordinate-system.code | example | Pattern: LOINC code 92822-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:coordinate-system.value[x] | extensible | LOINC LL5323-2http://loinc.org/vs/LL5323-2 | |
| Observation.component:ref-allele.code | example | Pattern: LOINC code 69547-8http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:alt-allele.code | example | Pattern: LOINC code 69551-0http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:coding-change-type.code | example | Pattern: LOINC code 48019-4http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:coding-change-type.value[x] | extensible | DNAChangeTypeVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/dna-change-type-vs | |
| Observation.component:genomic-source-class.code | example | Pattern: LOINC code 48002-0http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:genomic-source-class.value[x] | extensible | LOINC LL378-1http://loinc.org/vs/LL378-1 | |
| Observation.component:sample-allelic-frequency.code | example | Pattern: LOINC code 81258-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:sample-allelic-frequency.value[x].comparator | required | QuantityComparatorhttp://hl7.org/fhir/ValueSet/quantity-comparator|4.0.1from the FHIR Standard | |
| Observation.component:allelic-read-depth.code | example | Pattern: LOINC code 82121-5http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:allelic-state.code | example | Pattern: LOINC code 53034-5http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:allelic-state.value[x] | extensible | LOINC LL381-5http://loinc.org/vs/LL381-5 | |
| Observation.component:variant-inheritance.code | example | Pattern: variant-inheritancehttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:variant-inheritance.value[x] | extensible | VariantInheritanceVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/variant-inheritance-vs | |
| Observation.component:variation-code.code | example | Pattern: LOINC code 81252-9http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:variation-code.value[x] | example | ||
| Observation.component:chromosome-identifier.code | example | Pattern: LOINC code 48000-4http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:chromosome-identifier.value[x] | required | LOINC LL2938-0http://loinc.org/vs/LL2938-0 | |
| Observation.component:protein-hgvs.code | example | Pattern: LOINC code 48005-3http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:protein-hgvs.value[x] | required | HGVSVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/hgvs-vs | |
| Observation.component:amino-acid-change-type.code | example | Pattern: LOINC code 48006-1http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:amino-acid-change-type.value[x] | extensible | LOINC LL380-7http://loinc.org/vs/LL380-7 | |
| Observation.component:molecular-consequence.code | example | Pattern: molecular-consequencehttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:molecular-consequence.value[x] | extensible | MolecularConsequenceVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/molecular-consequence-vs | |
| Observation.component:copy-number.code | example | Pattern: LOINC code 82155-3http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:variant-confidence-status.code | example | Pattern: variant-confidence-statushttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:variant-confidence-status.value[x] | required | VariantConfidenceStatusVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/variant-confidence-status-vs |
| Name | Flags | Card. | Type | Description & Constraints | ||||
|---|---|---|---|---|---|---|---|---|
 | C | 0..* | Variant | Variant obs-6: dataAbsentReason SHALL only be present if Observation.value[x] is not present obs-7: If Observation.code is the same as an Observation.component.code then the value element associated with the code SHALL NOT be present | ||||
  | Σ | 0..1 | id | Logical id of this artifact | ||||
  | Σ | 0..1 | Meta | Metadata about the resource | ||||
  | ?!Σ | 0..1 | uri | A set of rules under which this content was created | ||||
  | 0..1 | code | Language of the resource content Binding: CommonLanguages (preferred): A human language.
| |||||
  | 0..1 | Narrative | Text summary of the resource, for human interpretation | |||||
  | 0..* | Resource | Contained, inline Resources | |||||
  | 0..* | Extension | Extension Slice: Unordered, Open by value:url | |||||
  | 0..1 | CodeableConcept | Secondary findings are genetic test results that provide information about variants in a gene unrelated to the primary purpose for the testing, most often discovered when [Whole Exome Sequencing (WES)](https://en.wikipedia.org/wiki/Exome_sequencing) or [Whole Genome Sequencing (WGS)](https://en.wikipedia.org/wiki/Whole_genome_sequencing) is performed. This extension should be used to denote when a genetic finding is being shared as a secondary finding, and ideally refer to a corresponding guideline or policy statement.
For more detail, please see:
https://ghr.nlm.nih.gov/primer/testing/secondaryfindings URL: http://hl7.org/fhir/StructureDefinition/observation-secondaryFinding Binding: GeneticObservationSecondaryFindings (extensible): Codes to denote a guideline or policy statement when a genetic test result is being shared as a secondary finding. | |||||
  | 0..1 | Reference(BodyStructure) | Target anatomic location or structure URL: http://hl7.org/fhir/StructureDefinition/bodySite | |||||
  | ?! | 0..* | Extension | Extensions that cannot be ignored | ||||
  | Σ | 0..* | Identifier | Business Identifier for observation | ||||
  | Σ | 0..* | Reference(CarePlan | DeviceRequest | ImmunizationRecommendation | MedicationRequest | NutritionOrder | ServiceRequest) | Fulfills plan, proposal or order | ||||
  | Σ | 0..* | Reference(MedicationAdministration | MedicationDispense | MedicationStatement | Procedure | Immunization | ImagingStudy) | Part of referenced event | ||||
  | ?!SΣ | 1..1 | code | registered | preliminary | final | amended + Binding: ObservationStatus (required): Codes providing the status of an observation. | ||||
  | S | 1..* | CodeableConcept | Classification of type of observation Slice: Unordered, Open by value:coding Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. | ||||
   | 1..1 | CodeableConcept | Classification of type of observation Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. Required Pattern: At least the following | |||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |||||
     | 0..1 | string | Version of the system - if relevant | |||||
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |||||
     | 0..1 | string | Representation defined by the system | |||||
     | 0..1 | boolean | If this coding was chosen directly by the user | |||||
    | 0..1 | string | Plain text representation of the concept | |||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations Slice: Unordered, Open by value:url | |||||
    | Σ | 1..1 | Coding | Code defined by a terminology system Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |||||
     | 0..1 | string | Version of the system - if relevant | |||||
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |||||
     | 0..1 | string | Representation defined by the system | |||||
     | 0..1 | boolean | If this coding was chosen directly by the user | |||||
    | Σ | 0..1 | string | Plain text representation of the concept | ||||
  | SΣ | 1..1 | CodeableConcept | 69548-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
   | 0..1 | string | Unique id for inter-element referencing | |||||
   | 0..* | Extension | Additional content defined by implementations | |||||
   | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
    | 0..1 | string | Version of the system - if relevant | |||||
    | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 69548-6 | |||||
    | 0..1 | string | Representation defined by the system | |||||
    | 0..1 | boolean | If this coding was chosen directly by the user | |||||
   | 0..1 | string | Plain text representation of the concept | |||||
  | SΣ | 0..1 | Reference(Cancer Patient Profile) | Who and/or what the observation is about | ||||
  | Σ | 0..* | Reference(Resource) | What the observation is about, when it is not about the subject of record | ||||
  | Σ | 0..1 | Reference(Encounter) | Healthcare event during which this observation is made | ||||
  | SΣ | 0..1 | Clinically relevant time/time-period for observation Slice: Unordered, Open by type:$this | |||||
   | dateTime | |||||||
   | Period | |||||||
   | Timing | |||||||
   | instant | |||||||
  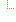 | SΣ | 0..1 | dateTime | Clinically relevant time/time-period for observation | ||||
  | Σ | 0..1 | instant | Date/Time this version was made available | ||||
  | Σ | 0..* | Reference(Practitioner | PractitionerRole | Organization | CareTeam | Patient | RelatedPerson) | Who is responsible for the observation | ||||
  | SΣC | 0..1 | CodeableConcept | Actual result Slice: Unordered, Closed by type:$this | ||||
  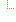 | ΣC | 0..1 | CodeableConcept | Indeterminate | No call | Present | Absent. Binding: LOINC Answer List LL1971-2 (required) | ||||
  | SC | 0..1 | CodeableConcept | Why the result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
  | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
  | 0..* | CodedAnnotation | Comments about the Observation that also contain a coded type | |||||
  | 0..1 | CodeableConcept | Observed body part Binding: SNOMEDCTBodyStructures (example): Codes describing anatomical locations. May include laterality. | |||||
  | S | 0..1 | CodeableConcept | Sequencing | SNP array | PCR | Computational analysis | ... Binding: LOINC Answer List LL4048-6 (extensible) | ||||
  | S | 0..1 | Reference(Human Specimen Profile) | Specimen used for this observation | ||||
  | 0..1 | Reference(Device | DeviceMetric) | (Measurement) Device | |||||
  | C | 0..* | BackboneElement | Not used in this profile obs-3: Must have at least a low or a high or text | ||||
   | 0..1 | string | Unique id for inter-element referencing | |||||
   | 0..* | Extension | Additional content defined by implementations | |||||
   | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
   | C | 0..1 | SimpleQuantity | Low Range, if relevant | ||||
   | C | 0..1 | SimpleQuantity | High Range, if relevant | ||||
   | 0..1 | CodeableConcept | Reference range qualifier Binding: ObservationReferenceRangeMeaningCodes (preferred): Code for the meaning of a reference range. | |||||
   | 0..* | CodeableConcept | Reference range population Binding: ObservationReferenceRangeAppliesToCodes (example): Codes identifying the population the reference range applies to. | |||||
   | 0..1 | Range | Applicable age range, if relevant | |||||
   | 0..1 | string | Text based reference range in an observation | |||||
  | Σ | 0..* | Reference(Observation | QuestionnaireResponse | MolecularSequence) | Not used in this profile | ||||
  | Σ | 0..* | Reference(DocumentReference | ImagingStudy | Media | QuestionnaireResponse | Observation | MolecularSequence) | Related measurements the observation is made from | ||||
  | SΣ | 0..* | BackboneElement | Component results Slice: Unordered, Open by pattern:code | ||||
   | Content/Rules for all slices | |||||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. | ||||
    | Σ | 0..1 | Actual component result | |||||
     | Quantity | |||||||
     | CodeableConcept | |||||||
     | string | |||||||
     | boolean | |||||||
     | integer | |||||||
     | Range | |||||||
     | Ratio | |||||||
     | SampledData | |||||||
     | time | |||||||
     | dateTime | |||||||
     | Period | |||||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Clinical Conclusion | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | conclusion-string Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: conclusion-string | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 0..1 | string | Summary conclusion (interpretation/impression) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..* | BackboneElement | Gene Studied | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 48018-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48018-6 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | The HGNC gene symbol is to be used as display text and the HGNC gene ID used as the code. If no HGNC code issued for this gene yet, NCBI gene IDs SHALL be used. Binding: HUGO Gene Nomenclature Committee Gene Names (HGNC) (extensible) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..* | BackboneElement | Cytogenetic (Chromosome) Location | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 48001-2 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48001-2 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | Example: 1q21.1 | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..* | BackboneElement | Human Reference Sequence Assembly | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 62374-4 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 62374-4 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 1..1 | CodeableConcept | GRCh37 | GRCh38 | ... Binding: LOINC Answer List LL1040-6 (extensible) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Coding (cDNA) Change - cHGVS | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 48004-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48004-6 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 1..1 | CodeableConcept | A valid HGVS-formatted 'c.' string, e.g. NM_005228.5:c.2369C>T. Binding: Human Genome Variation Society (HGVS) Nomenclature (required) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Genomic (gDNA) Change - gHGVS | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 81290-9 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81290-9 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | A valid HGVS-formatted 'g.' string, e.g. NC_000016.9:g.2124200_2138612dup Binding: Human Genome Variation Society (HGVS) Nomenclature (required) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Cytogenomic Nomenclature (ISCN) | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 81291-7 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81291-7 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Genomic Reference Sequence | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 48013-7 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48013-7 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 1..1 | CodeableConcept | Versioned genomic reference sequence identifier Binding: (unbound) (example): Multiple bindings acceptable (NCBI or LRG) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Reference Transcript | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 51958-7 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 51958-7 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 1..1 | CodeableConcept | Versioned transcript reference sequence identifier Binding: (unbound) (example): Multiple bindings acceptable (NCBI or LRG) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Exact Start-End | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 81254-5 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81254-5 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 0..1 | Range | Range in question. 'High' can be omitted for single nucleotide variants. | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Inner Start-End | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 81302-2 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81302-2 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 0..1 | Range | Imprecise variant inner-bounding range | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Outer Start-End | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 81301-4 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81301-4 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 0..1 | Range | Imprecise variant outer-bounding range | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Coordinate System | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 92822-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 92822-6 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 1..1 | CodeableConcept | 0-based interval counting | 0-based character counting | 1-based character counting Binding: LOINC Answer List LL5323-2 (extensible) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Genomic Ref Allele | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 69547-8 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 69547-8 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 0..1 | string | Normalized string per the VCF format. | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Genomic Alt Allele | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 69551-0 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 69551-0 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 0..1 | string | Normalized string per the VCF format. | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Coding DNA Change Type | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 48019-4 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48019-4 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | deletion | insertion | delins | SNV | copy_number_gain | copy_number_loss | ... (many) Binding: DNA Change Type (extensible): Concepts in sequence ontology under SO:0002072 | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Genomic Source Class | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 48002-0 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48002-0 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | Germline | Somatic | Fetal | Likely germline | Likely somatic | Likely fetal | Unknown genomic origin | De novo Binding: LOINC Answer List LL378-1 (extensible) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Sample Allelic Frequency | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 81258-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81258-6 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 0..1 | Quantity | Relative frequency in the sample | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations Slice: Unordered, Open by value:url | |||||
     | Σ | 0..1 | decimal | Numerical value (with implicit precision) | ||||
     | ?!Σ | 0..1 | code | < | <= | >= | > - how to understand the value Binding: QuantityComparator (required): How the Quantity should be understood and represented. | ||||
     | Σ | 0..1 | string | Unit representation | ||||
     | ΣC | 0..1 | uri | System that defines coded unit form Required Pattern: http://unitsofmeasure.org | ||||
     | Σ | 0..1 | code | Coded form of the unit | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Allelic Read Depth | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 82121-5 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 82121-5 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 0..1 | Quantity | Unfiltered count of supporting reads | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Allelic State | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 53034-5 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 53034-5 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | Heteroplasmic | Homoplasmic | Homozygous | Heterozygous | Hemizygous Binding: LOINC Answer List LL381-5 (extensible) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Variant Inheritance | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | variant-inheritance Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: variant-inheritance | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 1..1 | CodeableConcept | Maternal | Paternal | Unknown Binding: Variant Inheritances (extensible) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..* | BackboneElement | Variation Code | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 81252-9 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81252-9 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | ClinVar ID or similar Binding: (unbound) (example): Multiple bindings acceptable | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..* | BackboneElement | Chromosome Identifier | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 48000-4 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48000-4 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 1..1 | CodeableConcept | Chromosome 1 | Chromosome 2 | ... | Chromosome 22 | Chromosome X | Chromosome Y Binding: LOINC Answer List LL2938-0 (required) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Protein (Amino Acid) Change - pHGVS | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 48005-3 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48005-3 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | A valid HGVS-formatted 'p.' string, e.g. NP_000050.2:p.(Asn1836Lys) Binding: Human Genome Variation Society (HGVS) Nomenclature (required) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Amino Acid Change Type | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 48006-1 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48006-1 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | Wild type | Deletion | Duplication | Frameshift | Initiating Methionine | Insertion | Insertion and Deletion | Missense | Nonsense | Silent Binding: LOINC Answer List LL380-7 (extensible) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Molecular Consequence | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | molecular-consequence Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: molecular-consequence | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | stop_lost | stop_gained | inframe_insertion | frameshift_variant | ... (many) Binding: Molecular Consequence (extensible): Concepts in sequence ontology under SO:0001537. | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Genomic Structural Variant Copy Number | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 82155-3 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 82155-3 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣC | 0..1 | Quantity | Actual component result cnt-3: There SHALL be a code with a value of '1' if there is a value. If system is present, it SHALL be UCUM. If present, the value SHALL be a whole number. | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Variant Confidence Status | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | variant-confidence-status Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: variant-confidence-status | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 1..1 | CodeableConcept | High | Intermediate | Low Binding: Variant Confidence Status (required) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
 Documentation for this format Documentation for this format | ||||||||
| Path | Conformance | ValueSet / Code | URI | |||
| Observation.language | preferred | CommonLanguages
http://hl7.org/fhir/ValueSet/languagesfrom the FHIR Standard | ||||
| Observation.status | required | ObservationStatushttp://hl7.org/fhir/ValueSet/observation-status|4.0.1from the FHIR Standard | ||||
| Observation.category | preferred | ObservationCategoryCodeshttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | ||||
| Observation.category:labCategory | preferred | Pattern: laboratoryhttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | ||||
| Observation.code | example | Pattern: LOINC code 69548-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.value[x]:valueCodeableConcept | required | LOINC LL1971-2http://loinc.org/vs/LL1971-2 | ||||
| Observation.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.bodySite | example | SNOMEDCTBodyStructureshttp://hl7.org/fhir/ValueSet/body-sitefrom the FHIR Standard | ||||
| Observation.method | extensible | LOINC LL4048-6http://loinc.org/vs/LL4048-6 | ||||
| Observation.referenceRange.type | preferred | ObservationReferenceRangeMeaningCodeshttp://hl7.org/fhir/ValueSet/referencerange-meaningfrom the FHIR Standard | ||||
| Observation.referenceRange.appliesTo | example | ObservationReferenceRangeAppliesToCodeshttp://hl7.org/fhir/ValueSet/referencerange-appliestofrom the FHIR Standard | ||||
| Observation.component.code | example | LOINCCodes (a valid code from LOINC)http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:conclusion-string.code | example | Pattern: conclusion-stringhttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:conclusion-string.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:conclusion-string.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:gene-studied.code | example | Pattern: LOINC code 48018-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:gene-studied.value[x] | extensible | HGNCVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/hgnc-vs | ||||
| Observation.component:gene-studied.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:gene-studied.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:cytogenetic-location.code | example | Pattern: LOINC code 48001-2http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:cytogenetic-location.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:cytogenetic-location.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:reference-sequence-assembly.code | example | Pattern: LOINC code 62374-4http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:reference-sequence-assembly.value[x] | extensible | LOINC LL1040-6http://loinc.org/vs/LL1040-6 | ||||
| Observation.component:reference-sequence-assembly.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:reference-sequence-assembly.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:coding-hgvs.code | example | Pattern: LOINC code 48004-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:coding-hgvs.value[x] | required | HGVSVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/hgvs-vs | ||||
| Observation.component:coding-hgvs.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:coding-hgvs.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:genomic-hgvs.code | example | Pattern: LOINC code 81290-9http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:genomic-hgvs.value[x] | required | HGVSVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/hgvs-vs | ||||
| Observation.component:genomic-hgvs.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:genomic-hgvs.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:cytogenomic-nomenclature.code | example | Pattern: LOINC code 81291-7http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:cytogenomic-nomenclature.value[x] | example | |||||
| Observation.component:cytogenomic-nomenclature.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:cytogenomic-nomenclature.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:genomic-ref-seq.code | example | Pattern: LOINC code 48013-7http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:genomic-ref-seq.value[x] | example | |||||
| Observation.component:genomic-ref-seq.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:genomic-ref-seq.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:transcript-ref-seq.code | example | Pattern: LOINC code 51958-7http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:transcript-ref-seq.value[x] | example | |||||
| Observation.component:transcript-ref-seq.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:transcript-ref-seq.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:exact-start-end.code | example | Pattern: LOINC code 81254-5http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:exact-start-end.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:exact-start-end.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:inner-start-end.code | example | Pattern: LOINC code 81302-2http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:inner-start-end.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:inner-start-end.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:outer-start-end.code | example | Pattern: LOINC code 81301-4http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:outer-start-end.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:outer-start-end.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:coordinate-system.code | example | Pattern: LOINC code 92822-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:coordinate-system.value[x] | extensible | LOINC LL5323-2http://loinc.org/vs/LL5323-2 | ||||
| Observation.component:coordinate-system.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:coordinate-system.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:ref-allele.code | example | Pattern: LOINC code 69547-8http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:ref-allele.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:ref-allele.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:alt-allele.code | example | Pattern: LOINC code 69551-0http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:alt-allele.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:alt-allele.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:coding-change-type.code | example | Pattern: LOINC code 48019-4http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:coding-change-type.value[x] | extensible | DNAChangeTypeVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/dna-change-type-vs | ||||
| Observation.component:coding-change-type.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:coding-change-type.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:genomic-source-class.code | example | Pattern: LOINC code 48002-0http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:genomic-source-class.value[x] | extensible | LOINC LL378-1http://loinc.org/vs/LL378-1 | ||||
| Observation.component:genomic-source-class.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:genomic-source-class.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:sample-allelic-frequency.code | example | Pattern: LOINC code 81258-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:sample-allelic-frequency.value[x].comparator | required | QuantityComparatorhttp://hl7.org/fhir/ValueSet/quantity-comparator|4.0.1from the FHIR Standard | ||||
| Observation.component:sample-allelic-frequency.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:sample-allelic-frequency.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:allelic-read-depth.code | example | Pattern: LOINC code 82121-5http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:allelic-read-depth.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:allelic-read-depth.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:allelic-state.code | example | Pattern: LOINC code 53034-5http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:allelic-state.value[x] | extensible | LOINC LL381-5http://loinc.org/vs/LL381-5 | ||||
| Observation.component:allelic-state.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:allelic-state.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:variant-inheritance.code | example | Pattern: variant-inheritancehttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:variant-inheritance.value[x] | extensible | VariantInheritanceVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/variant-inheritance-vs | ||||
| Observation.component:variant-inheritance.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:variant-inheritance.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:variation-code.code | example | Pattern: LOINC code 81252-9http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:variation-code.value[x] | example | |||||
| Observation.component:variation-code.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:variation-code.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:chromosome-identifier.code | example | Pattern: LOINC code 48000-4http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:chromosome-identifier.value[x] | required | LOINC LL2938-0http://loinc.org/vs/LL2938-0 | ||||
| Observation.component:chromosome-identifier.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:chromosome-identifier.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:protein-hgvs.code | example | Pattern: LOINC code 48005-3http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:protein-hgvs.value[x] | required | HGVSVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/hgvs-vs | ||||
| Observation.component:protein-hgvs.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:protein-hgvs.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:amino-acid-change-type.code | example | Pattern: LOINC code 48006-1http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:amino-acid-change-type.value[x] | extensible | LOINC LL380-7http://loinc.org/vs/LL380-7 | ||||
| Observation.component:amino-acid-change-type.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:amino-acid-change-type.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:molecular-consequence.code | example | Pattern: molecular-consequencehttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:molecular-consequence.value[x] | extensible | MolecularConsequenceVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/molecular-consequence-vs | ||||
| Observation.component:molecular-consequence.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:molecular-consequence.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:copy-number.code | example | Pattern: LOINC code 82155-3http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:copy-number.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:copy-number.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:variant-confidence-status.code | example | Pattern: variant-confidence-statushttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:variant-confidence-status.value[x] | required | VariantConfidenceStatusVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/variant-confidence-status-vs | ||||
| Observation.component:variant-confidence-status.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:variant-confidence-status.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard |
This structure is derived from Variant
Summary
Must-Support: 50 elements
Structures
This structure refers to these other structures:
Slices
This structure defines the following Slices:
Maturity: 1
Differential View
This structure is derived from Variant
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | 0..* | Variant | Variant | |
  | S | 1..1 | code | registered | preliminary | final | amended + |
  | S | 1..* | CodeableConcept | Classification of type of observation |
  | 1..1 | CodeableConcept | Classification of type of observation Required Pattern: At least the following | |
   | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
    | 1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |
    | 1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |
   | 1..1 | Coding | Code defined by a terminology system | |
  | S | 1..1 | CodeableConcept | 69548-6 |
  | S | 0..1 | Reference(Cancer Patient Profile) | Who and/or what the observation is about |
  | S | 0..1 | dateTime, Period, Timing, instant | Clinically relevant time/time-period for observation Slice: Unordered, Open by type:$this |
   | S | 0..1 | dateTime | Clinically relevant time/time-period for observation |
  | S | 0..1 | CodeableConcept | Actual result |
  | S | 0..1 | CodeableConcept | Why the result is missing |
  | S | 0..1 | CodeableConcept | Sequencing | SNP array | PCR | Computational analysis | ... |
  | S | 0..1 | Reference(Human Specimen Profile) | Specimen used for this observation |
  | 0..* | BackboneElement | Not used in this profile | |
  | 0..* | Reference(Observation | QuestionnaireResponse | MolecularSequence) | Not used in this profile | |
  | S | 0..* | BackboneElement | Component results |
  | S | 0..* | BackboneElement | Gene Studied |
   | S | 1..1 | CodeableConcept | 48018-6 |
   | S | 1..1 | CodeableConcept | The HGNC gene symbol is to be used as display text and the HGNC gene ID used as the code. If no HGNC code issued for this gene yet, NCBI gene IDs SHALL be used. |
  | S | 0..* | BackboneElement | Cytogenetic (Chromosome) Location |
   | S | 1..1 | CodeableConcept | 48001-2 |
   | S | 1..1 | CodeableConcept | Example: 1q21.1 |
  | S | 0..1 | BackboneElement | Genomic (gDNA) Change - gHGVS |
   | S | 1..1 | CodeableConcept | 81290-9 |
   | S | 1..1 | CodeableConcept | A valid HGVS-formatted 'g.' string, e.g. NC_000016.9:g.2124200_2138612dup |
  | S | 0..1 | BackboneElement | Cytogenomic Nomenclature (ISCN) |
   | S | 1..1 | CodeableConcept | 81291-7 |
   | S | 1..1 | CodeableConcept | Actual component result |
  | S | 0..1 | BackboneElement | Coding DNA Change Type |
   | S | 1..1 | CodeableConcept | 48019-4 |
   | S | 1..1 | CodeableConcept | deletion | insertion | delins | SNV | copy_number_gain | copy_number_loss | ... (many) |
  | S | 0..1 | BackboneElement | Genomic Source Class |
   | S | 1..1 | CodeableConcept | 48002-0 |
   | S | 1..1 | CodeableConcept | Germline | Somatic | Fetal | Likely germline | Likely somatic | Likely fetal | Unknown genomic origin | De novo |
  | S | 0..1 | BackboneElement | Sample Allelic Frequency |
   | S | 1..1 | CodeableConcept | 81258-6 |
   | S | 0..1 | Quantity | Relative frequency in the sample |
  | S | 0..1 | BackboneElement | Allelic State |
   | S | 1..1 | CodeableConcept | 53034-5 |
   | S | 1..1 | CodeableConcept | Heteroplasmic | Homoplasmic | Homozygous | Heterozygous | Hemizygous |
  | S | 0..* | BackboneElement | Variation Code |
   | S | 1..1 | CodeableConcept | 81252-9 |
   | S | 1..1 | CodeableConcept | ClinVar ID or similar |
  | S | 0..1 | BackboneElement | Protein (Amino Acid) Change - pHGVS |
   | S | 1..1 | CodeableConcept | 48005-3 |
   | S | 1..1 | CodeableConcept | A valid HGVS-formatted 'p.' string, e.g. NP_000050.2:p.(Asn1836Lys) |
  | S | 0..1 | BackboneElement | Amino Acid Change Type |
   | S | 1..1 | CodeableConcept | 48006-1 |
   | S | 1..1 | CodeableConcept | Wild type | Deletion | Duplication | Frameshift | Initiating Methionine | Insertion | Insertion and Deletion | Missense | Nonsense | Silent |
  | S | 0..1 | BackboneElement | Molecular Consequence |
   | S | 1..1 | CodeableConcept | molecular-consequence |
   | S | 1..1 | CodeableConcept | stop_lost | stop_gained | inframe_insertion | frameshift_variant | ... (many) |
  | S | 0..1 | BackboneElement | Genomic Structural Variant Copy Number |
   | S | 1..1 | CodeableConcept | 82155-3 |
   | S | 0..1 | Quantity | Actual component result |
 Documentation for this format Documentation for this format | ||||
Key Elements View
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | C | 0..* | Variant | Variant obs-6: dataAbsentReason SHALL only be present if Observation.value[x] is not present obs-7: If Observation.code is the same as an Observation.component.code then the value element associated with the code SHALL NOT be present |
  | ?!Σ | 0..1 | uri | A set of rules under which this content was created |
  | 0..1 | CodeableConcept | Secondary findings are genetic test results that provide information about variants in a gene unrelated to the primary purpose for the testing, most often discovered when [Whole Exome Sequencing (WES)](https://en.wikipedia.org/wiki/Exome_sequencing) or [Whole Genome Sequencing (WGS)](https://en.wikipedia.org/wiki/Whole_genome_sequencing) is performed. This extension should be used to denote when a genetic finding is being shared as a secondary finding, and ideally refer to a corresponding guideline or policy statement.
For more detail, please see:
https://ghr.nlm.nih.gov/primer/testing/secondaryfindings URL: http://hl7.org/fhir/StructureDefinition/observation-secondaryFinding Binding: GeneticObservationSecondaryFindings (extensible): Codes to denote a guideline or policy statement when a genetic test result is being shared as a secondary finding. | |
  | 0..1 | Reference(BodyStructure) | Target anatomic location or structure URL: http://hl7.org/fhir/StructureDefinition/bodySite | |
  | ?! | 0..* | Extension | Extensions that cannot be ignored |
  | ?!SΣ | 1..1 | code | registered | preliminary | final | amended + Binding: ObservationStatus (required): Codes providing the status of an observation. |
  | S | 1..* | CodeableConcept | Classification of type of observation Slice: Unordered, Open by value:coding Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. |
   | 1..1 | CodeableConcept | Classification of type of observation Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. Required Pattern: At least the following | |
    | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |
    | Σ | 1..1 | Coding | Code defined by a terminology system Required Pattern: At least the following |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |
  | SΣ | 1..1 | CodeableConcept | 69548-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
   | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
    | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
    | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 69548-6 | |
  | SΣ | 0..1 | Reference(Cancer Patient Profile) | Who and/or what the observation is about |
  | SΣ | 0..1 | Clinically relevant time/time-period for observation Slice: Unordered, Open by type:$this | |
   | dateTime | |||
   | Period | |||
   | Timing | |||
   | instant | |||
   | SΣ | 0..1 | dateTime | Clinically relevant time/time-period for observation |
  | SΣC | 0..1 | CodeableConcept | Actual result Slice: Unordered, Closed by type:$this |
   | ΣC | 0..1 | CodeableConcept | Indeterminate | No call | Present | Absent. Binding: LOINC Answer List LL1971-2 (required) |
  | SC | 0..1 | CodeableConcept | Why the result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
  | S | 0..1 | CodeableConcept | Sequencing | SNP array | PCR | Computational analysis | ... Binding: LOINC Answer List LL4048-6 (extensible) |
  | S | 0..1 | Reference(Human Specimen Profile) | Specimen used for this observation |
  | C | 0..* | BackboneElement | Not used in this profile obs-3: Must have at least a low or a high or text |
   | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
  | Σ | 0..* | Reference(Observation | QuestionnaireResponse | MolecularSequence) | Not used in this profile |
  | SΣ | 0..* | BackboneElement | Component results Slice: Unordered, Open by pattern:code |
   | Content/Rules for all slices | |||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. |
   | Σ | 0..1 | BackboneElement | Clinical Conclusion |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | conclusion-string Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: conclusion-string | |
   | SΣ | 0..* | BackboneElement | Gene Studied |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 48018-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48018-6 | |
    | SΣ | 1..1 | CodeableConcept | The HGNC gene symbol is to be used as display text and the HGNC gene ID used as the code. If no HGNC code issued for this gene yet, NCBI gene IDs SHALL be used. Binding: HUGO Gene Nomenclature Committee Gene Names (HGNC) (extensible) |
   | SΣ | 0..* | BackboneElement | Cytogenetic (Chromosome) Location |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 48001-2 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48001-2 | |
    | SΣ | 1..1 | CodeableConcept | Example: 1q21.1 |
   | Σ | 0..* | BackboneElement | Human Reference Sequence Assembly |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 62374-4 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 62374-4 | |
    | Σ | 1..1 | CodeableConcept | GRCh37 | GRCh38 | ... Binding: LOINC Answer List LL1040-6 (extensible) |
   | Σ | 0..1 | BackboneElement | Coding (cDNA) Change - cHGVS |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 48004-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48004-6 | |
    | Σ | 1..1 | CodeableConcept | A valid HGVS-formatted 'c.' string, e.g. NM_005228.5:c.2369C>T. Binding: Human Genome Variation Society (HGVS) Nomenclature (required) |
   | SΣ | 0..1 | BackboneElement | Genomic (gDNA) Change - gHGVS |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 81290-9 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81290-9 | |
    | SΣ | 1..1 | CodeableConcept | A valid HGVS-formatted 'g.' string, e.g. NC_000016.9:g.2124200_2138612dup Binding: Human Genome Variation Society (HGVS) Nomenclature (required) |
   | SΣ | 0..1 | BackboneElement | Cytogenomic Nomenclature (ISCN) |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 81291-7 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81291-7 | |
    | SΣ | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined |
   | Σ | 0..1 | BackboneElement | Genomic Reference Sequence |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 48013-7 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48013-7 | |
    | Σ | 1..1 | CodeableConcept | Versioned genomic reference sequence identifier Binding: (unbound) (example): Multiple bindings acceptable (NCBI or LRG) |
   | Σ | 0..1 | BackboneElement | Reference Transcript |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 51958-7 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 51958-7 | |
    | Σ | 1..1 | CodeableConcept | Versioned transcript reference sequence identifier Binding: (unbound) (example): Multiple bindings acceptable (NCBI or LRG) |
   | Σ | 0..1 | BackboneElement | Exact Start-End |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 81254-5 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81254-5 | |
   | Σ | 0..1 | BackboneElement | Inner Start-End |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 81302-2 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81302-2 | |
   | Σ | 0..1 | BackboneElement | Outer Start-End |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 81301-4 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81301-4 | |
   | Σ | 0..1 | BackboneElement | Coordinate System |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 92822-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 92822-6 | |
    | Σ | 1..1 | CodeableConcept | 0-based interval counting | 0-based character counting | 1-based character counting Binding: LOINC Answer List LL5323-2 (extensible) |
   | Σ | 0..1 | BackboneElement | Genomic Ref Allele |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 69547-8 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 69547-8 | |
   | Σ | 0..1 | BackboneElement | Genomic Alt Allele |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 69551-0 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 69551-0 | |
   | SΣ | 0..1 | BackboneElement | Coding DNA Change Type |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 48019-4 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48019-4 | |
    | SΣ | 1..1 | CodeableConcept | deletion | insertion | delins | SNV | copy_number_gain | copy_number_loss | ... (many) Binding: DNA Change Type (extensible): Concepts in sequence ontology under SO:0002072 |
   | SΣ | 0..1 | BackboneElement | Genomic Source Class |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 48002-0 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48002-0 | |
    | SΣ | 1..1 | CodeableConcept | Germline | Somatic | Fetal | Likely germline | Likely somatic | Likely fetal | Unknown genomic origin | De novo Binding: LOINC Answer List LL378-1 (extensible) |
   | SΣ | 0..1 | BackboneElement | Sample Allelic Frequency |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 81258-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81258-6 | |
    | SΣ | 0..1 | Quantity | Relative frequency in the sample |
     | ?!Σ | 0..1 | code | < | <= | >= | > - how to understand the value Binding: QuantityComparator (required): How the Quantity should be understood and represented. |
   | Σ | 0..1 | BackboneElement | Allelic Read Depth |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 82121-5 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 82121-5 | |
   | SΣ | 0..1 | BackboneElement | Allelic State |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 53034-5 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 53034-5 | |
    | SΣ | 1..1 | CodeableConcept | Heteroplasmic | Homoplasmic | Homozygous | Heterozygous | Hemizygous Binding: LOINC Answer List LL381-5 (extensible) |
   | Σ | 0..1 | BackboneElement | Variant Inheritance |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | variant-inheritance Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: variant-inheritance | |
    | Σ | 1..1 | CodeableConcept | Maternal | Paternal | Unknown Binding: Variant Inheritances (extensible) |
   | SΣ | 0..* | BackboneElement | Variation Code |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 81252-9 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81252-9 | |
    | SΣ | 1..1 | CodeableConcept | ClinVar ID or similar Binding: (unbound) (example): Multiple bindings acceptable |
   | Σ | 0..* | BackboneElement | Chromosome Identifier |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | 48000-4 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48000-4 | |
    | Σ | 1..1 | CodeableConcept | Chromosome 1 | Chromosome 2 | ... | Chromosome 22 | Chromosome X | Chromosome Y Binding: LOINC Answer List LL2938-0 (required) |
   | SΣ | 0..1 | BackboneElement | Protein (Amino Acid) Change - pHGVS |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 48005-3 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48005-3 | |
    | SΣ | 1..1 | CodeableConcept | A valid HGVS-formatted 'p.' string, e.g. NP_000050.2:p.(Asn1836Lys) Binding: Human Genome Variation Society (HGVS) Nomenclature (required) |
   | SΣ | 0..1 | BackboneElement | Amino Acid Change Type |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 48006-1 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48006-1 | |
    | SΣ | 1..1 | CodeableConcept | Wild type | Deletion | Duplication | Frameshift | Initiating Methionine | Insertion | Insertion and Deletion | Missense | Nonsense | Silent Binding: LOINC Answer List LL380-7 (extensible) |
   | SΣ | 0..1 | BackboneElement | Molecular Consequence |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | molecular-consequence Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: molecular-consequence | |
    | SΣ | 1..1 | CodeableConcept | stop_lost | stop_gained | inframe_insertion | frameshift_variant | ... (many) Binding: Molecular Consequence (extensible): Concepts in sequence ontology under SO:0001537. |
   | SΣ | 0..1 | BackboneElement | Genomic Structural Variant Copy Number |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | SΣ | 1..1 | CodeableConcept | 82155-3 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 82155-3 | |
    | SΣC | 0..1 | Quantity | Actual component result cnt-3: There SHALL be a code with a value of '1' if there is a value. If system is present, it SHALL be UCUM. If present, the value SHALL be a whole number. |
   | Σ | 0..1 | BackboneElement | Variant Confidence Status |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | variant-confidence-status Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: variant-confidence-status | |
    | Σ | 1..1 | CodeableConcept | High | Intermediate | Low Binding: Variant Confidence Status (required) |
 Documentation for this format Documentation for this format | ||||
| Path | Conformance | ValueSet / Code | URI |
| Observation.status | required | ObservationStatushttp://hl7.org/fhir/ValueSet/observation-status|4.0.1from the FHIR Standard | |
| Observation.category | preferred | ObservationCategoryCodeshttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | |
| Observation.category:labCategory | preferred | Pattern: laboratoryhttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | |
| Observation.code | example | Pattern: LOINC code 69548-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.value[x]:valueCodeableConcept | required | LOINC LL1971-2http://loinc.org/vs/LL1971-2 | |
| Observation.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | |
| Observation.method | extensible | LOINC LL4048-6http://loinc.org/vs/LL4048-6 | |
| Observation.component.code | example | LOINCCodes (a valid code from LOINC)http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:conclusion-string.code | example | Pattern: conclusion-stringhttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:gene-studied.code | example | Pattern: LOINC code 48018-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:gene-studied.value[x] | extensible | HGNCVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/hgnc-vs | |
| Observation.component:cytogenetic-location.code | example | Pattern: LOINC code 48001-2http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:reference-sequence-assembly.code | example | Pattern: LOINC code 62374-4http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:reference-sequence-assembly.value[x] | extensible | LOINC LL1040-6http://loinc.org/vs/LL1040-6 | |
| Observation.component:coding-hgvs.code | example | Pattern: LOINC code 48004-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:coding-hgvs.value[x] | required | HGVSVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/hgvs-vs | |
| Observation.component:genomic-hgvs.code | example | Pattern: LOINC code 81290-9http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:genomic-hgvs.value[x] | required | HGVSVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/hgvs-vs | |
| Observation.component:cytogenomic-nomenclature.code | example | Pattern: LOINC code 81291-7http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:cytogenomic-nomenclature.value[x] | example | ||
| Observation.component:genomic-ref-seq.code | example | Pattern: LOINC code 48013-7http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:genomic-ref-seq.value[x] | example | ||
| Observation.component:transcript-ref-seq.code | example | Pattern: LOINC code 51958-7http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:transcript-ref-seq.value[x] | example | ||
| Observation.component:exact-start-end.code | example | Pattern: LOINC code 81254-5http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:inner-start-end.code | example | Pattern: LOINC code 81302-2http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:outer-start-end.code | example | Pattern: LOINC code 81301-4http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:coordinate-system.code | example | Pattern: LOINC code 92822-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:coordinate-system.value[x] | extensible | LOINC LL5323-2http://loinc.org/vs/LL5323-2 | |
| Observation.component:ref-allele.code | example | Pattern: LOINC code 69547-8http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:alt-allele.code | example | Pattern: LOINC code 69551-0http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:coding-change-type.code | example | Pattern: LOINC code 48019-4http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:coding-change-type.value[x] | extensible | DNAChangeTypeVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/dna-change-type-vs | |
| Observation.component:genomic-source-class.code | example | Pattern: LOINC code 48002-0http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:genomic-source-class.value[x] | extensible | LOINC LL378-1http://loinc.org/vs/LL378-1 | |
| Observation.component:sample-allelic-frequency.code | example | Pattern: LOINC code 81258-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:sample-allelic-frequency.value[x].comparator | required | QuantityComparatorhttp://hl7.org/fhir/ValueSet/quantity-comparator|4.0.1from the FHIR Standard | |
| Observation.component:allelic-read-depth.code | example | Pattern: LOINC code 82121-5http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:allelic-state.code | example | Pattern: LOINC code 53034-5http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:allelic-state.value[x] | extensible | LOINC LL381-5http://loinc.org/vs/LL381-5 | |
| Observation.component:variant-inheritance.code | example | Pattern: variant-inheritancehttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:variant-inheritance.value[x] | extensible | VariantInheritanceVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/variant-inheritance-vs | |
| Observation.component:variation-code.code | example | Pattern: LOINC code 81252-9http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:variation-code.value[x] | example | ||
| Observation.component:chromosome-identifier.code | example | Pattern: LOINC code 48000-4http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:chromosome-identifier.value[x] | required | LOINC LL2938-0http://loinc.org/vs/LL2938-0 | |
| Observation.component:protein-hgvs.code | example | Pattern: LOINC code 48005-3http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:protein-hgvs.value[x] | required | HGVSVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/hgvs-vs | |
| Observation.component:amino-acid-change-type.code | example | Pattern: LOINC code 48006-1http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:amino-acid-change-type.value[x] | extensible | LOINC LL380-7http://loinc.org/vs/LL380-7 | |
| Observation.component:molecular-consequence.code | example | Pattern: molecular-consequencehttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:molecular-consequence.value[x] | extensible | MolecularConsequenceVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/molecular-consequence-vs | |
| Observation.component:copy-number.code | example | Pattern: LOINC code 82155-3http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:variant-confidence-status.code | example | Pattern: variant-confidence-statushttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:variant-confidence-status.value[x] | required | VariantConfidenceStatusVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/variant-confidence-status-vs |
Snapshot View
| Name | Flags | Card. | Type | Description & Constraints | ||||
|---|---|---|---|---|---|---|---|---|
 | C | 0..* | Variant | Variant obs-6: dataAbsentReason SHALL only be present if Observation.value[x] is not present obs-7: If Observation.code is the same as an Observation.component.code then the value element associated with the code SHALL NOT be present | ||||
  | Σ | 0..1 | id | Logical id of this artifact | ||||
  | Σ | 0..1 | Meta | Metadata about the resource | ||||
  | ?!Σ | 0..1 | uri | A set of rules under which this content was created | ||||
  | 0..1 | code | Language of the resource content Binding: CommonLanguages (preferred): A human language.
| |||||
  | 0..1 | Narrative | Text summary of the resource, for human interpretation | |||||
  | 0..* | Resource | Contained, inline Resources | |||||
  | 0..* | Extension | Extension Slice: Unordered, Open by value:url | |||||
  | 0..1 | CodeableConcept | Secondary findings are genetic test results that provide information about variants in a gene unrelated to the primary purpose for the testing, most often discovered when [Whole Exome Sequencing (WES)](https://en.wikipedia.org/wiki/Exome_sequencing) or [Whole Genome Sequencing (WGS)](https://en.wikipedia.org/wiki/Whole_genome_sequencing) is performed. This extension should be used to denote when a genetic finding is being shared as a secondary finding, and ideally refer to a corresponding guideline or policy statement.
For more detail, please see:
https://ghr.nlm.nih.gov/primer/testing/secondaryfindings URL: http://hl7.org/fhir/StructureDefinition/observation-secondaryFinding Binding: GeneticObservationSecondaryFindings (extensible): Codes to denote a guideline or policy statement when a genetic test result is being shared as a secondary finding. | |||||
  | 0..1 | Reference(BodyStructure) | Target anatomic location or structure URL: http://hl7.org/fhir/StructureDefinition/bodySite | |||||
  | ?! | 0..* | Extension | Extensions that cannot be ignored | ||||
  | Σ | 0..* | Identifier | Business Identifier for observation | ||||
  | Σ | 0..* | Reference(CarePlan | DeviceRequest | ImmunizationRecommendation | MedicationRequest | NutritionOrder | ServiceRequest) | Fulfills plan, proposal or order | ||||
  | Σ | 0..* | Reference(MedicationAdministration | MedicationDispense | MedicationStatement | Procedure | Immunization | ImagingStudy) | Part of referenced event | ||||
  | ?!SΣ | 1..1 | code | registered | preliminary | final | amended + Binding: ObservationStatus (required): Codes providing the status of an observation. | ||||
  | S | 1..* | CodeableConcept | Classification of type of observation Slice: Unordered, Open by value:coding Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. | ||||
   | 1..1 | CodeableConcept | Classification of type of observation Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. Required Pattern: At least the following | |||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |||||
     | 0..1 | string | Version of the system - if relevant | |||||
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |||||
     | 0..1 | string | Representation defined by the system | |||||
     | 0..1 | boolean | If this coding was chosen directly by the user | |||||
    | 0..1 | string | Plain text representation of the concept | |||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations Slice: Unordered, Open by value:url | |||||
    | Σ | 1..1 | Coding | Code defined by a terminology system Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |||||
     | 0..1 | string | Version of the system - if relevant | |||||
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |||||
     | 0..1 | string | Representation defined by the system | |||||
     | 0..1 | boolean | If this coding was chosen directly by the user | |||||
    | Σ | 0..1 | string | Plain text representation of the concept | ||||
  | SΣ | 1..1 | CodeableConcept | 69548-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
   | 0..1 | string | Unique id for inter-element referencing | |||||
   | 0..* | Extension | Additional content defined by implementations | |||||
   | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
    | 0..1 | string | Version of the system - if relevant | |||||
    | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 69548-6 | |||||
    | 0..1 | string | Representation defined by the system | |||||
    | 0..1 | boolean | If this coding was chosen directly by the user | |||||
   | 0..1 | string | Plain text representation of the concept | |||||
  | SΣ | 0..1 | Reference(Cancer Patient Profile) | Who and/or what the observation is about | ||||
  | Σ | 0..* | Reference(Resource) | What the observation is about, when it is not about the subject of record | ||||
  | Σ | 0..1 | Reference(Encounter) | Healthcare event during which this observation is made | ||||
  | SΣ | 0..1 | Clinically relevant time/time-period for observation Slice: Unordered, Open by type:$this | |||||
   | dateTime | |||||||
   | Period | |||||||
   | Timing | |||||||
   | instant | |||||||
   | SΣ | 0..1 | dateTime | Clinically relevant time/time-period for observation | ||||
  | Σ | 0..1 | instant | Date/Time this version was made available | ||||
  | Σ | 0..* | Reference(Practitioner | PractitionerRole | Organization | CareTeam | Patient | RelatedPerson) | Who is responsible for the observation | ||||
  | SΣC | 0..1 | CodeableConcept | Actual result Slice: Unordered, Closed by type:$this | ||||
   | ΣC | 0..1 | CodeableConcept | Indeterminate | No call | Present | Absent. Binding: LOINC Answer List LL1971-2 (required) | ||||
  | SC | 0..1 | CodeableConcept | Why the result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
  | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
  | 0..* | CodedAnnotation | Comments about the Observation that also contain a coded type | |||||
  | 0..1 | CodeableConcept | Observed body part Binding: SNOMEDCTBodyStructures (example): Codes describing anatomical locations. May include laterality. | |||||
  | S | 0..1 | CodeableConcept | Sequencing | SNP array | PCR | Computational analysis | ... Binding: LOINC Answer List LL4048-6 (extensible) | ||||
  | S | 0..1 | Reference(Human Specimen Profile) | Specimen used for this observation | ||||
  | 0..1 | Reference(Device | DeviceMetric) | (Measurement) Device | |||||
  | C | 0..* | BackboneElement | Not used in this profile obs-3: Must have at least a low or a high or text | ||||
   | 0..1 | string | Unique id for inter-element referencing | |||||
   | 0..* | Extension | Additional content defined by implementations | |||||
   | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
   | C | 0..1 | SimpleQuantity | Low Range, if relevant | ||||
   | C | 0..1 | SimpleQuantity | High Range, if relevant | ||||
   | 0..1 | CodeableConcept | Reference range qualifier Binding: ObservationReferenceRangeMeaningCodes (preferred): Code for the meaning of a reference range. | |||||
   | 0..* | CodeableConcept | Reference range population Binding: ObservationReferenceRangeAppliesToCodes (example): Codes identifying the population the reference range applies to. | |||||
   | 0..1 | Range | Applicable age range, if relevant | |||||
   | 0..1 | string | Text based reference range in an observation | |||||
  | Σ | 0..* | Reference(Observation | QuestionnaireResponse | MolecularSequence) | Not used in this profile | ||||
  | Σ | 0..* | Reference(DocumentReference | ImagingStudy | Media | QuestionnaireResponse | Observation | MolecularSequence) | Related measurements the observation is made from | ||||
  | SΣ | 0..* | BackboneElement | Component results Slice: Unordered, Open by pattern:code | ||||
   | Content/Rules for all slices | |||||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. | ||||
    | Σ | 0..1 | Actual component result | |||||
     | Quantity | |||||||
     | CodeableConcept | |||||||
     | string | |||||||
     | boolean | |||||||
     | integer | |||||||
     | Range | |||||||
     | Ratio | |||||||
     | SampledData | |||||||
     | time | |||||||
     | dateTime | |||||||
     | Period | |||||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Clinical Conclusion | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | conclusion-string Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: conclusion-string | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 0..1 | string | Summary conclusion (interpretation/impression) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..* | BackboneElement | Gene Studied | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 48018-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48018-6 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | The HGNC gene symbol is to be used as display text and the HGNC gene ID used as the code. If no HGNC code issued for this gene yet, NCBI gene IDs SHALL be used. Binding: HUGO Gene Nomenclature Committee Gene Names (HGNC) (extensible) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..* | BackboneElement | Cytogenetic (Chromosome) Location | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 48001-2 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48001-2 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | Example: 1q21.1 | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..* | BackboneElement | Human Reference Sequence Assembly | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 62374-4 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 62374-4 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 1..1 | CodeableConcept | GRCh37 | GRCh38 | ... Binding: LOINC Answer List LL1040-6 (extensible) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Coding (cDNA) Change - cHGVS | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 48004-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48004-6 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 1..1 | CodeableConcept | A valid HGVS-formatted 'c.' string, e.g. NM_005228.5:c.2369C>T. Binding: Human Genome Variation Society (HGVS) Nomenclature (required) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Genomic (gDNA) Change - gHGVS | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 81290-9 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81290-9 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | A valid HGVS-formatted 'g.' string, e.g. NC_000016.9:g.2124200_2138612dup Binding: Human Genome Variation Society (HGVS) Nomenclature (required) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Cytogenomic Nomenclature (ISCN) | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 81291-7 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81291-7 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Genomic Reference Sequence | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 48013-7 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48013-7 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 1..1 | CodeableConcept | Versioned genomic reference sequence identifier Binding: (unbound) (example): Multiple bindings acceptable (NCBI or LRG) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Reference Transcript | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 51958-7 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 51958-7 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 1..1 | CodeableConcept | Versioned transcript reference sequence identifier Binding: (unbound) (example): Multiple bindings acceptable (NCBI or LRG) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Exact Start-End | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 81254-5 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81254-5 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 0..1 | Range | Range in question. 'High' can be omitted for single nucleotide variants. | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Inner Start-End | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 81302-2 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81302-2 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 0..1 | Range | Imprecise variant inner-bounding range | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Outer Start-End | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 81301-4 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81301-4 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 0..1 | Range | Imprecise variant outer-bounding range | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Coordinate System | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 92822-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 92822-6 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 1..1 | CodeableConcept | 0-based interval counting | 0-based character counting | 1-based character counting Binding: LOINC Answer List LL5323-2 (extensible) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Genomic Ref Allele | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 69547-8 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 69547-8 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 0..1 | string | Normalized string per the VCF format. | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Genomic Alt Allele | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 69551-0 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 69551-0 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 0..1 | string | Normalized string per the VCF format. | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Coding DNA Change Type | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 48019-4 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48019-4 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | deletion | insertion | delins | SNV | copy_number_gain | copy_number_loss | ... (many) Binding: DNA Change Type (extensible): Concepts in sequence ontology under SO:0002072 | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Genomic Source Class | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 48002-0 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48002-0 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | Germline | Somatic | Fetal | Likely germline | Likely somatic | Likely fetal | Unknown genomic origin | De novo Binding: LOINC Answer List LL378-1 (extensible) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Sample Allelic Frequency | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 81258-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81258-6 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 0..1 | Quantity | Relative frequency in the sample | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations Slice: Unordered, Open by value:url | |||||
     | Σ | 0..1 | decimal | Numerical value (with implicit precision) | ||||
     | ?!Σ | 0..1 | code | < | <= | >= | > - how to understand the value Binding: QuantityComparator (required): How the Quantity should be understood and represented. | ||||
     | Σ | 0..1 | string | Unit representation | ||||
     | ΣC | 0..1 | uri | System that defines coded unit form Required Pattern: http://unitsofmeasure.org | ||||
     | Σ | 0..1 | code | Coded form of the unit | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Allelic Read Depth | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 82121-5 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 82121-5 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 0..1 | Quantity | Unfiltered count of supporting reads | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Allelic State | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 53034-5 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 53034-5 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | Heteroplasmic | Homoplasmic | Homozygous | Heterozygous | Hemizygous Binding: LOINC Answer List LL381-5 (extensible) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Variant Inheritance | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | variant-inheritance Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: variant-inheritance | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 1..1 | CodeableConcept | Maternal | Paternal | Unknown Binding: Variant Inheritances (extensible) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..* | BackboneElement | Variation Code | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 81252-9 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81252-9 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | ClinVar ID or similar Binding: (unbound) (example): Multiple bindings acceptable | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..* | BackboneElement | Chromosome Identifier | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | 48000-4 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48000-4 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 1..1 | CodeableConcept | Chromosome 1 | Chromosome 2 | ... | Chromosome 22 | Chromosome X | Chromosome Y Binding: LOINC Answer List LL2938-0 (required) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Protein (Amino Acid) Change - pHGVS | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 48005-3 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48005-3 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | A valid HGVS-formatted 'p.' string, e.g. NP_000050.2:p.(Asn1836Lys) Binding: Human Genome Variation Society (HGVS) Nomenclature (required) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Amino Acid Change Type | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 48006-1 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 48006-1 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | Wild type | Deletion | Duplication | Frameshift | Initiating Methionine | Insertion | Insertion and Deletion | Missense | Nonsense | Silent Binding: LOINC Answer List LL380-7 (extensible) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Molecular Consequence | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | molecular-consequence Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: molecular-consequence | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣ | 1..1 | CodeableConcept | stop_lost | stop_gained | inframe_insertion | frameshift_variant | ... (many) Binding: Molecular Consequence (extensible): Concepts in sequence ontology under SO:0001537. | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | SΣ | 0..1 | BackboneElement | Genomic Structural Variant Copy Number | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | SΣ | 1..1 | CodeableConcept | 82155-3 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 82155-3 | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | SΣC | 0..1 | Quantity | Actual component result cnt-3: There SHALL be a code with a value of '1' if there is a value. If system is present, it SHALL be UCUM. If present, the value SHALL be a whole number. | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   | Σ | 0..1 | BackboneElement | Variant Confidence Status | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    | Σ | 1..1 | CodeableConcept | variant-confidence-status Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     | 0..1 | string | Unique id for inter-element referencing | |||||
     | 0..* | Extension | Additional content defined by implementations | |||||
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      | 0..1 | string | Unique id for inter-element referencing | |||||
      | 0..* | Extension | Additional content defined by implementations | |||||
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |||||
      | 0..1 | string | Version of the system - if relevant | |||||
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: variant-confidence-status | |||||
      | 0..1 | string | Representation defined by the system | |||||
      | 0..1 | boolean | If this coding was chosen directly by the user | |||||
     | 0..1 | string | Plain text representation of the concept | |||||
    | Σ | 1..1 | CodeableConcept | High | Intermediate | Low Binding: Variant Confidence Status (required) | ||||
    | C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
 Documentation for this format Documentation for this format | ||||||||
| Path | Conformance | ValueSet / Code | URI | |||
| Observation.language | preferred | CommonLanguages
http://hl7.org/fhir/ValueSet/languagesfrom the FHIR Standard | ||||
| Observation.status | required | ObservationStatushttp://hl7.org/fhir/ValueSet/observation-status|4.0.1from the FHIR Standard | ||||
| Observation.category | preferred | ObservationCategoryCodeshttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | ||||
| Observation.category:labCategory | preferred | Pattern: laboratoryhttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | ||||
| Observation.code | example | Pattern: LOINC code 69548-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.value[x]:valueCodeableConcept | required | LOINC LL1971-2http://loinc.org/vs/LL1971-2 | ||||
| Observation.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.bodySite | example | SNOMEDCTBodyStructureshttp://hl7.org/fhir/ValueSet/body-sitefrom the FHIR Standard | ||||
| Observation.method | extensible | LOINC LL4048-6http://loinc.org/vs/LL4048-6 | ||||
| Observation.referenceRange.type | preferred | ObservationReferenceRangeMeaningCodeshttp://hl7.org/fhir/ValueSet/referencerange-meaningfrom the FHIR Standard | ||||
| Observation.referenceRange.appliesTo | example | ObservationReferenceRangeAppliesToCodeshttp://hl7.org/fhir/ValueSet/referencerange-appliestofrom the FHIR Standard | ||||
| Observation.component.code | example | LOINCCodes (a valid code from LOINC)http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:conclusion-string.code | example | Pattern: conclusion-stringhttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:conclusion-string.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:conclusion-string.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:gene-studied.code | example | Pattern: LOINC code 48018-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:gene-studied.value[x] | extensible | HGNCVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/hgnc-vs | ||||
| Observation.component:gene-studied.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:gene-studied.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:cytogenetic-location.code | example | Pattern: LOINC code 48001-2http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:cytogenetic-location.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:cytogenetic-location.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:reference-sequence-assembly.code | example | Pattern: LOINC code 62374-4http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:reference-sequence-assembly.value[x] | extensible | LOINC LL1040-6http://loinc.org/vs/LL1040-6 | ||||
| Observation.component:reference-sequence-assembly.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:reference-sequence-assembly.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:coding-hgvs.code | example | Pattern: LOINC code 48004-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:coding-hgvs.value[x] | required | HGVSVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/hgvs-vs | ||||
| Observation.component:coding-hgvs.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:coding-hgvs.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:genomic-hgvs.code | example | Pattern: LOINC code 81290-9http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:genomic-hgvs.value[x] | required | HGVSVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/hgvs-vs | ||||
| Observation.component:genomic-hgvs.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:genomic-hgvs.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:cytogenomic-nomenclature.code | example | Pattern: LOINC code 81291-7http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:cytogenomic-nomenclature.value[x] | example | |||||
| Observation.component:cytogenomic-nomenclature.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:cytogenomic-nomenclature.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:genomic-ref-seq.code | example | Pattern: LOINC code 48013-7http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:genomic-ref-seq.value[x] | example | |||||
| Observation.component:genomic-ref-seq.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:genomic-ref-seq.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:transcript-ref-seq.code | example | Pattern: LOINC code 51958-7http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:transcript-ref-seq.value[x] | example | |||||
| Observation.component:transcript-ref-seq.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:transcript-ref-seq.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:exact-start-end.code | example | Pattern: LOINC code 81254-5http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:exact-start-end.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:exact-start-end.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:inner-start-end.code | example | Pattern: LOINC code 81302-2http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:inner-start-end.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:inner-start-end.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:outer-start-end.code | example | Pattern: LOINC code 81301-4http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:outer-start-end.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:outer-start-end.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:coordinate-system.code | example | Pattern: LOINC code 92822-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:coordinate-system.value[x] | extensible | LOINC LL5323-2http://loinc.org/vs/LL5323-2 | ||||
| Observation.component:coordinate-system.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:coordinate-system.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:ref-allele.code | example | Pattern: LOINC code 69547-8http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:ref-allele.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:ref-allele.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:alt-allele.code | example | Pattern: LOINC code 69551-0http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:alt-allele.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:alt-allele.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:coding-change-type.code | example | Pattern: LOINC code 48019-4http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:coding-change-type.value[x] | extensible | DNAChangeTypeVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/dna-change-type-vs | ||||
| Observation.component:coding-change-type.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:coding-change-type.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:genomic-source-class.code | example | Pattern: LOINC code 48002-0http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:genomic-source-class.value[x] | extensible | LOINC LL378-1http://loinc.org/vs/LL378-1 | ||||
| Observation.component:genomic-source-class.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:genomic-source-class.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:sample-allelic-frequency.code | example | Pattern: LOINC code 81258-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:sample-allelic-frequency.value[x].comparator | required | QuantityComparatorhttp://hl7.org/fhir/ValueSet/quantity-comparator|4.0.1from the FHIR Standard | ||||
| Observation.component:sample-allelic-frequency.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:sample-allelic-frequency.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:allelic-read-depth.code | example | Pattern: LOINC code 82121-5http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:allelic-read-depth.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:allelic-read-depth.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:allelic-state.code | example | Pattern: LOINC code 53034-5http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:allelic-state.value[x] | extensible | LOINC LL381-5http://loinc.org/vs/LL381-5 | ||||
| Observation.component:allelic-state.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:allelic-state.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:variant-inheritance.code | example | Pattern: variant-inheritancehttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:variant-inheritance.value[x] | extensible | VariantInheritanceVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/variant-inheritance-vs | ||||
| Observation.component:variant-inheritance.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:variant-inheritance.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:variation-code.code | example | Pattern: LOINC code 81252-9http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:variation-code.value[x] | example | |||||
| Observation.component:variation-code.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:variation-code.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:chromosome-identifier.code | example | Pattern: LOINC code 48000-4http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:chromosome-identifier.value[x] | required | LOINC LL2938-0http://loinc.org/vs/LL2938-0 | ||||
| Observation.component:chromosome-identifier.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:chromosome-identifier.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:protein-hgvs.code | example | Pattern: LOINC code 48005-3http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:protein-hgvs.value[x] | required | HGVSVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/hgvs-vs | ||||
| Observation.component:protein-hgvs.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:protein-hgvs.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:amino-acid-change-type.code | example | Pattern: LOINC code 48006-1http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:amino-acid-change-type.value[x] | extensible | LOINC LL380-7http://loinc.org/vs/LL380-7 | ||||
| Observation.component:amino-acid-change-type.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:amino-acid-change-type.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:molecular-consequence.code | example | Pattern: molecular-consequencehttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:molecular-consequence.value[x] | extensible | MolecularConsequenceVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/molecular-consequence-vs | ||||
| Observation.component:molecular-consequence.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:molecular-consequence.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:copy-number.code | example | Pattern: LOINC code 82155-3http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:copy-number.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:copy-number.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:variant-confidence-status.code | example | Pattern: variant-confidence-statushttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:variant-confidence-status.value[x] | required | VariantConfidenceStatusVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/variant-confidence-status-vs | ||||
| Observation.component:variant-confidence-status.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:variant-confidence-status.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard |
This structure is derived from Variant
Summary
Must-Support: 50 elements
Structures
This structure refers to these other structures:
Slices
This structure defines the following Slices:
Maturity: 1
Other representations of profile: CSV, Excel, Schematron