Requirements Federated Learning and mUlti-party computation Techniques for prostatE cancer
0.1.0 - ci-build
![]()
Requirements Federated Learning and mUlti-party computation Techniques for prostatE cancer
0.1.0 - ci-build
![]()
Requirements Federated Learning and mUlti-party computation Techniques for prostatE cancer, published by HL7 Europe. This guide is not an authorized publication; it is the continuous build for version 0.1.0 built by the FHIR (HL7® FHIR® Standard) CI Build. This version is based on the current content of https://github.com/hl7-eu/flute-requirements/ and changes regularly. See the Directory of published versions
FLUTE specific requirements are outlined based on discussions with stakeholders participating in the case studies, in particular representatives of technical partners participating in WP5 (Quibim) and medical researchers from the three participating hospitals CHUL, IRST and VHIR, which also act as data owners. Data owners are aware of ICT aspects, data protection and other requirements for the data owner nodes. The part also includes GDPR considerations for FLUTE pilot studies and the platform.
The study is an analytical observational, retrospective, and multicenter study based on 3 different European institutions: Hospital Universitari Vall d’Hebrón (HUVH, Spain), Instituto Romagnolo per lo Study dei Tumori (IRST, Italy) and Centre Hospitalier Universitaire de Liège (CHU, Belgium). The retrospective cohorts consist of men with clinical suspicion of PCa based on a PSA > 3.0 ng/ml and/or abnormal DRE, in whom the 7 clinical variables used in the Barcelona Predictive Model of csPCa are retrospectively collected. Each patient underwent a mpMRI/bpMRI reported with the Prostate Imaging-Report and Data System (PI-RADS), and a subsequent systematic and/or targeted (aimed at PI-RADS ≥3 lesions) prostate biopsy during the first year after the MRI.
From each MRI, a prospectively radiomics analysis is performed to extract the quantitative imaging biomarkers from the automatic segmentation of anatomic prostate regions/suspicious lesions with QP-Prostate, an AI-based software developed by QUIBIM.
The imaging biomarkers will be added to the 7 clinical variables of BCN-RC to create a predictive model for the prediction of csPCA in patients with clinical suspicion of PCa
Number of subjects:
Patient inclusion criteria
Men with clinical suspicion of PCa based on a PSA > 3.0 ng/ml and/or abnormal DRE, in whom a mpMRI/bpMRI is performed and a subsequent systematic and/or targeted prostate biopsy is done during the first year following the MRI. Lesions detected in mpMRI/bpMRI have to be reported using the Prostate Imaging-Report and Data System (PI-RADS) in version 2.0 or higher. Prostate biopsies are systematic and targeted in cases of PI-RADS ≥3 lesions.
Patient exclusion criteria
Patients without MRI images prior to the biopsy or with images obtained earlier than one year before the biopsy
List of variables The variables to be extracted from each cohort and to be included in the model are defined as follows: Endpoint variable: csPCa, defined as a PCa in prostate biopsy with an International Society of Urologic Pathology (ISUP) grade group (GG) 2 or higher. Independent variables:
Types of variables that will be included in the model:
| Variable | Description | Type of data | Data format | Source system |
|---|---|---|---|---|
| AGE | Age at the biopsy | numeric | integer | Clinical History |
| FH | PCa family history | categorical | 0: No; 1: Yes | Clinical History |
| TB | Type of biopsy | categorical | 0: initial; 2: repeated | Clinical History/Procedure report |
| PSA | PSA | numeric | Numeric with 1 decimal (ng/ml) | Clinical History/Lab data |
| DRE | Rectal examination | categorical | 0: normal; 1: suspicious | Clinical History |
| VP | MRI-prostate volume | numeric | Numeric with 1 decimal (cc) | MRI report |
| PIRADS | PI-RADS v.2.0 or 2.1 | categorical | 1 to 5 | MRI report |
For a successful launch of QP-Prostate®, the study must include the T2-weighted MR sequence and the DWI. DCE sequences are optional, but should be uploaded in case a multiparametric study has been acquired.
The analysis inclusion criteria for the T2W, DWI and DCE sequences are based on PI-RADS® v2.1 recommendations. However, QP-Prostate® software is also able to analyzed cases not compliant with PI-RADS ® v2.1, but should follow the acceptable criteria described in the Annex “Requirements QP-Prostate.pdf”. In the mentioned Annex, the requirements for running QP-Prostate® are described.
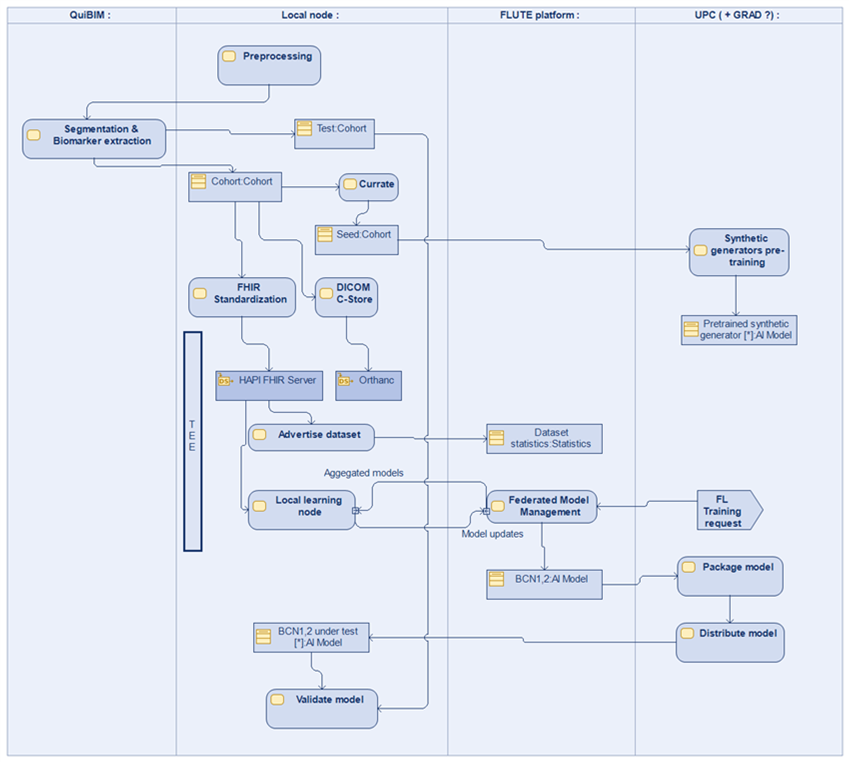
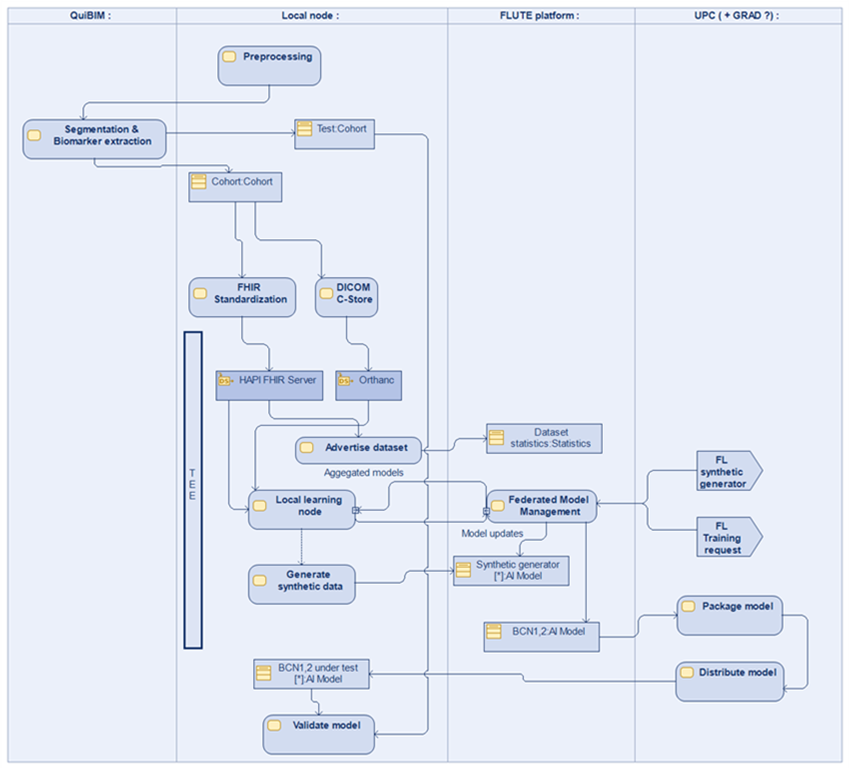
The variables detailed above will be extracted from EHR and PACS at CHUL, HUVH and IRST. The final databases will be stored in the FLUTE local nodes installed at each clinical site. Each FLUTE node can participate in the federated FLUTE research network. The FLUTE platform uses cryptographic methods like homomorphic encryption, SMPC, TEE and differential privacy to aggregate statistics about the cohort without leaking sensitive data outside the local node. Iteratively, the FLUTE node can train machine learning algorithms to fit the given datasets.
Within the FLUTE platform, each study is associated to a privacy budget. The FLUTE platform will ensure that the privacy budget is not exhausted. The FLUTE platform provides a graphical use interface to researchers to: a) discover and select the datasets registered at each FLUTE node; b) obtain descriptive statistics about the datasets; c) fit statistical and AI models to the datasets. The later part is provided using Jupyter notebooks integrated with the FLUTE functionalities.
First phase The first iteration will
Second phase The second iteration will be the development of the model(s)
The project aims to build a local data node at the data custodian premises. This node will be populated with data coming from different IT sub-systems of the data custodian or external (EHR, PACS, QP-Prostate, etc.). To demonstrate the capability of the platform to train a model on a specific cohort, a superset of the clinical cohort will be uploaded. E.g., Reduced sample datasets with the 7 clinical variables and associated images will be uploaded to the FLUTE node.
GDPR compliance requirements for case studies General Data Protection Regulation (GDPR) regulates the use of personal data and provides for specific requirements for fair and lawful processing of such data. The case studies will require collection and use of retrospective medical data provided by Hospital Universitari Vall d’Hebrón (VHIR, Spain), Instituto Romagnolo per lo Study dei Tumori (IRST, Italy) and Centre Hospitalier Universitaire de Liège (CHU, Belgium). Data relating to health is considered personal data under GDPR and requires an elevated level of data protection as special category of data (often referred to as ‘sensitive data’). Below we provide an overview of the legal considerations for the use of such data in the case studies.
The GDPR empowers Member States to impose derogations and exceptions in respect of GDPR obligations for particular processing activities. This allowance is extended to processing for scientific research purposes. Article 89 of the GDPR governs processing for scientific, historical or statistical purposes. Data Controllers are permitted to process data for these specific purposes where appropriate safeguards are implemented in accordance with Article 89(1). In this specific context, Article 89(2) of the GDPR allows Member States to establish further derogations from data subjects right referred to in Article 15, 16, 18 and 21, hence the legal basis for the processing will depend on the particular Data Provider:
In accordance with Article 89 §1 of the GDPR (echoed by Article 197 of the Belgian Law 30 July 2018 on the Protection of Individuals with regard to the Processing of Personal Data) the data controller using personal data for scientific research purposes must implement safeguards, which ensure technical and organizational measures to ensure the respect for data minimization. In particular in the following ways:
In relation to the FLUTE project, for data contributed by CHUL and VHIR, the research objectives cannot be achieved using anonymous data. For CHUL, it is required to keep the code to allow patients to exercise their right to object and to troubleshoot ETL processes from the different hospital IT systems involved. For VHIR, anonymous data would be insufficient because images will be processed outside the hospital by QUIBIM and afterwards the results obtained need to be linked to the clinical records. Therefore, personal identifiers are coded when data is extracted from the hospital clinical IT system into the FLUTE platform.
For IRST, data will be predominantly anonymized, subject to few example cases which may need to be pseudonymized. In the latter case, patient consent will be obtained. The following activities could be performed at IRST iteratively:
Moreover, the FLUTE project starts from the idea that sensitive data must not leave the premises of the data owners (hospitals). Thus, once the data is prepared by the hospital, it resides on a server (called ‘data owner node’) protected by the hospital’s own infrastructure. This server via FLUTE platform then exchanges encrypted messages with other data owners to collaboratively compute aggregates (e.g., averages of attributes or gradients, or other statistics) in such a way that under the security assumptions: (i) no sensitive information can be revealed from these exchanged messages;(ii) only a privatized version of the aggregate/model/statistic/etc. the data owners agreed in advance to compute can be revealed.
The specific exceptions to this rule, required to achieve the purposes of the project, shall be defined by the project.
The Consortium Agreement of the FLUTE project outlines the rights and obligation of the partners. A data protection impact analysis will be conducted with the hospitals’ DPOs to identify whether additional agreements are needed for data processing. Supporting data sharing agreements will be drafted and executed prior to data sharing.
GDPR impact on the functional requirements of the Platform In this section we build on the legal requirements which identified in Section 5.3 of the Trumpet deliverable 1.1. and provide additional comments translating the specified principles into functional requirements applicable to the Platform.
Article 5 of the GDPR provides for accountability by stipulating that the controller shall be responsible for, and be able to demonstrate compliance with the data protection principles of “lawfulness, fairness and transparency”, “purpose limitation”, “data minimization”, “accuracy”, “storage limitation” and security (“integrity and confidentiality”)”. Accountability means that entities responsible for the processing of data must be identified, and that appropriate controls (such as logs) are available to ensure that any problems can be attributed to the correct entity.
According to the principle of integrity and confidentiality (also referred to as “data security”, as elaborated in Article 32 GDPR), data must be protected by appropriate technical and organizational measures to ensure its confidentiality, integrity and availability. Data security refers to the layer of security in an information system that is devoted to adding protections to the data in the system itself and controlling the access to the data through identity and access management. FLUTE needs to implement security requirements that are appropriate to the sensitivity of the information at and that take into account the system and its components as well as the data accessible through the system and the identified risks. In particular, the risks may include:
Given the above risks, in the context of FLUTE, examples of the measures may include: